Matthew R Allegretti Week 10
From OpenWetWare
Jump to navigationJump to search
Week 10 Assignment
Purpose
To familiarize ourselves with protein sequencing and analysis tools. To understand how changes in amino acids and therefore changes in protein structure affect the function of gp120.
Methods and Results
The previous steps were performed in Week 9
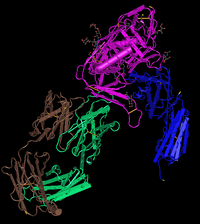
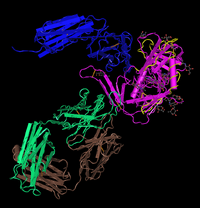
- These structure files can be opened with the Cn3D software site that is installed on the computers in the lab (this software is free, so you can download it and use it at home, too.) Alternately, you may choose to use the Star Biochem program to do this portion of your work. Answer the following:
- Find the N-terminus and C-terminus of each polypeptide tertiary structure.
- Locate all the secondary structure elements. Do these match the predictions made by the PredictProtein server?
- By comparing PredictProtein data and the structure model, the predictions made do match with the secondary structure at those locations.
- Locate the V3 region and figure out the location of the Markham et al. (1998) sequences in the structure.
- Perform a multiple sequence alignment on the protein sequences that you and your partner are analyzing for your project using the Biology Workbench.
- Are there more or fewer differences between the sequences when you look at the DNA sequences versus the protein sequences?
- There are more differences looking at the DNA than looking at the protein sequences.
- How do you account for this?
- Some base pair changes can result in synonymous mutations that do not effect the protein that is coded for within the reading frame.
- Are there more or fewer differences between the sequences when you look at the DNA sequences versus the protein sequences?
- Once you have oriented yourself, analyze whether the amino acid changes that you see in the multiple sequence alignment would affect the 3D structure and explain why you think this.
- Instead of using Cn3D or StarBiochem to do this, you could use the program ConSurf.
- While the region of the structure that would be affected by the changes in amino acids is limited to a 95 amino acid region, I believe that many of these changes would in fact affect the structure of the protein in this region. Many of the amino acid substitutions resulted in drastically different amino acid type or size. Few of the substitutions replaced one amino acid with a different one of a similar size or group.
- Instead of using Cn3D or StarBiochem to do this, you could use the program ConSurf.
Data and Files
Conclusion
- The purpose of this lab was to familiarize ourselves with protein sequencing tools, and to understand how changes in amino acids and therefore changes in protein structure affect the function of gp120. By performing the exercises, I was exposed to a variety of protein sequencing and analyzing tools, and have gained a greater understanding of how to examine the structure of a protein. Through our research project and presentation, we will examine the effects that amino acid substitutions cause within the gp120 protein and how those changes affect its functional success.
Research Project
Subject 4
- 3 Clones
- 13 Clones
Subject 9
- 5 Clones
- 11 Clones
Subject 11
- 7 Clones
- 9 Clones
Subject 14
- 6 Clones
- 10 Clones
- 64 Total Clones
Variability
- We observed the positions at which amino acid substitutions occurred, and variability can be measured either by:
- How many different amino acids are present at each position
- How many clones are affected by substitutions at that position
Method 1
- Single variability: 1 amino acid among all clones, but different from reference sequence
- Low variability group: 2 different amino acids present at each position among clones
- High variability: More than 2 different amino acids present at each position among clones
- Single variability
- Positions 275, 283, 289, 301, 305, 308, 339, 343, 346, 358, 361
- Low variability
- Positions 290, 299, 300, 302, 311, 313, 314, 321, 322, 324, 333, 335, 337, 345
- 321 is a technicality as some clone sequences did not have an amino acid for that position
- Positions 290, 299, 300, 302, 311, 313, 314, 321, 322, 324, 333, 335, 337, 345
- High variability
- 304, 306, 320, 327, 334, 353
- 320 is a technicality as some clone sequences did not have an amino acid for that position
- 304, 306, 320, 327, 334, 353
Data and Files
- Media:Matthew R Allegretti Markham S4V1 sequences.docx
- Media:Matthew R Allegretti Markham S4V4 sequences.docx
- Media:Matthew R Allegretti Markham S9V1 sequences.docx
- Media:Matthew R Allegretti Markham S9V4 sequences.docx
- Media:Matthew R Allegretti Markham S11V1 sequences.docx
- Media:Matthew R Allegretti Markham S11V4 sequences.docx
- Media:Matthew R Allegretti Markham S14V1 sequences.docx
- Media:Matthew R Allegretti Markham S14V4 sequences.docx
- Presentation
Acknowledgments
- Anu Varshneya
- We communicated electronically and collaborated for our presentation.
- Will Fuchs and Matthew Oki
- Collaborated with Will and Matt on our methods of comparing protein sequences.
While I worked with the people noted above, this individual journal entry was completed by me and not copied from another source.
- Dr. Dahlquist
Methods section pulled heavily from Dr. Dahlquist's Week 10 instructions
References
Useful links
- Stem Cell Overview
- Template
- Student Page
- Week 1 Instructions
- Journal Week 1
- Week 2 Instructions
- Week 2
- Journal Week 2
- Week 3 Instructions
- Week 3
- Journal Week 3
- Week 4 Instructions
- Week 4
- Journal Week 4
- Week 5 Instructions
- Week 5
- Journal Week 5
- Week 6 Instructions
- Week 6
- Journal Week 6
- Week 7 Instructions
- Journal Week 7
- Week 8 Instructions
- Week 8
- Journal Week 8
- Week 9 Instructions
- Week 9
- Journal Week 9
- Week 10 Instructions
- Week 10
- Journal Week 10
- Week 11 Instructions
- Week 11
- Journal Week 11
- Week 12 Instructions
- Week 14 Instructions
- Week 14 Assignment Part 1 and 2
- Journal Week 14
- Week 15 Instructions
- Week 15
- Journal Week 15
- My user page