Koch Lab:Publications
| Home | Research | Lab Members | Publications | Protocols | Contact | Funding |
| Principles | Data | Notebooks | Links | Meetings | Presentations | Inventory |
Kinesin / Microtubule Molecular Motor System
Effects of casein surface passivation in kinesin-1 gliding motility assay
Maloney A, Herskowitz LJ, Koch SJ (2011) Effects of Surface Passivation on Gliding Motility Assays. PLoS ONE 6(6): e19522. http://dx.doi.org/10.1371/journal.pone.0019522
Andy Maloney studies the effects of alpha, beta, kappa, and whole casein in the kinesin-1 gliding motility assay. Automated speed analysis software by Larry Herskowitz.
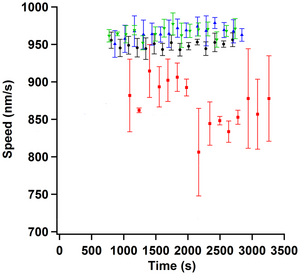
Figure: Black circles are alpha casein passivation, red squares are beta casein passivation, green up pointing triangles are whole casein passivation, and blue down pointing triangles are mixed casein passivation. Each data point is the mean from three different samples, taken at approximately the same assay time. Error bars represent the standard error of the mean. Alpha casein had the most consistent average speed measurements at 949±4 nm/s. Whole casein and mixed casein averaged to 966±7 nm/s and 966±6 nm/s respectively. Beta casein averaged to 870±30 nm/s.
A discrete-state model for kinesin-1 with rate constants modulated by neck linker tension (under review, PLoS ONE)
Koch, Steven and Herskowitz, Lawrence. A Discrete State Model for Kinesin-1 with Rate Constants Modulated by Neck Linker Tension. Available from Nature Precedings http://hdl.handle.net/10101/npre.2010.5038.1 (2010)
Effect of water isotope (deuterium or oxygen-18) on the kinesin-1 gliding motility assay (in preparation)
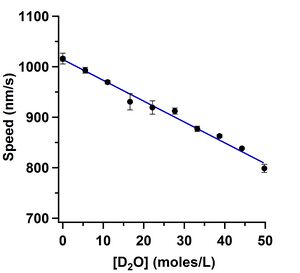
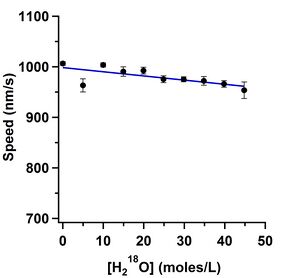
Andy Maloney took painstakingly clean data on how D2O and oxygen-18 water affect the gliding speed in the kinesin-1 gliding motility assay. We are preparing this data for submission to PLoS ONE. We are doing further investigation to understand what the mechanism is and whether it is an interesting way to probe this and other biomolecular systems. The effect is strikingly similar to the change in viscosity of the solvents, but we have not yet proven that is the mechanism.
See Andy Maloney's dissertation: http://www.openwetware.org/wiki/User:Andy_Maloney/Water_isotope_effects_on_kinesin_and_microtubules
Open Source Kinesin-1 Stochastic Simulation and Gliding Assay Tracking Software (pre-print only)
Herskowitz, Lawrence, Salvagno, Anthony, Josey, Brian, Maloney, Andy, and Koch, Steven. Open Source Kinesin-1 Stochastic Simulation and Gliding Assay Tracking Software. Available from Nature Precedings http://dx.doi.org/10.1038/npre.2010.4468.1 (2010)
Larry Herskowitz wrote this software to test his microtubule image tracking. It's a nice LabVIEW application for generating a series of images that simulate microtubules fractionally-labeled with dye molecules. You can design a route for the microtubules to follow. We are happy to provide the source code and help you get it running on your own system. We decided that it was not worth peer reviewers' time to submit this for formal publication, as we did not anticipate any benefits besides padding our resumes. So, we will leave it at the preprint stage on Nature Precedings.
In George Bachand's lab with Susan Rivera, fungal kinesin from Thermomyces lanuginosus
Rivera SB, Koch SJ, Bauer JM, Edwards JM, Bachand GD. 2007. "Temperature dependent properties of a kinesin-3 motor protein from Thermomyces lanuginosus." Fungal Genetics and Biology 44:1170-1179. PMID 17398126
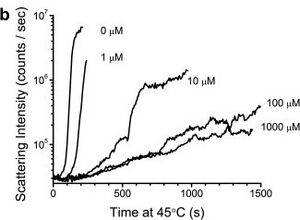
Figure: Dynamic light scattering data showing the inhibition by ATP of heat-induced aggregation of kinesin from Thermomyces lanuginosus.
Probing Protein-DNA Interactions by Unzipping DNA with Optical Tweezers
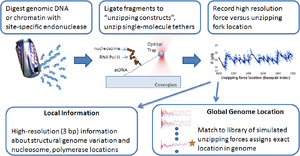
Our initial proof-of-principle publication in the Wang Lab at Cornell
Koch SJ, Shundrovsky A, Jantzen BC, Wang MD. Probing protein-DNA interactions by unzipping a single DNA double helix. Biophys J. 2002 Aug;83(2):1098-105. PMID 12124289
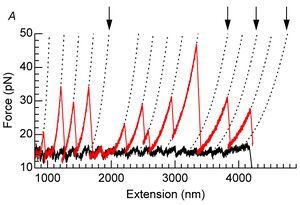
Our follow-on paper showing that unbinding forces can be analyzed nicely with Evan Evans' Dynamic Force Spectrosocpy (DFS) model
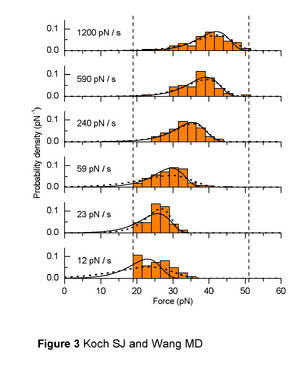
Koch SJ, Wang MD. Dynamic force spectroscopy of protein-DNA interactions by unzipping DNA. Phys Rev Lett. 2003 Jul 11;91(2):028103. PMID 12906513
- Buried in this paper is the "loading rate clamp" that we used and which greatly simplifies data analysis as well as provides much cleaner data. Also, our maximum likelihood method for data analysis is better than the typical method of fitting Gaussians to histograms, but this was also buried in footnotes. It's been while since published, but the Koch lab would like to publish the details of these methods, as they would be very helpful to others doing DFS.
DNA in porous nanochannels
Initial passive transport of DNA in porous nanochannels fabricated from silica nanoparticles

We have been consulting with the Brueck and Lopez labs at UNM on a project for transporting DNA in nanochannels where the walls are formed from silica nanoparticles are are thus porous on the nanoscale. Initial work of Deying Xia and Thomas Gamble were recently published in Nanoletters:
Xia Deying, Gamble Thomas C., Mendoza Edgar A., Koch Steven J., He Xiang, Lopez Gabriel P., and Brueck S. R. J. "DNA Transport in Hierarchically-Structured Colloidal-Nanoparticle Porous-Wall Nanochannels." Nano Lett., 8 (6) 1610 - 1618, 2008
MEMS Force Sensor for Biophysics
Work done with Gayle Thayer, Alex Corwin, Maarten de Boer at Sandia, finished up after Koch moved to UNM, Physics & CHTM
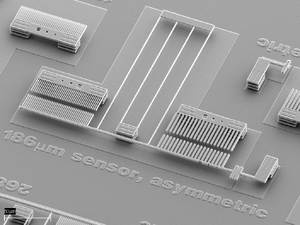
Koch SJ, Thayer GE, Corwin AD, de Boer MP. Micromachined piconewton force sensor for biophysics investigations. Appl. Phys. Let. 2006 Oct 23;89(17):173901 (PDF)
Book Chapters
Collaboration Using Open Notebook Science in Academia
Bradley J-C, Lang A SID, Koch SJ, Neylon C, "Collaboration Using Open Notebook Science in Academia" (2011). Chapter in Collaborative Computational Technologies for Biomedical Research, edited by Ekins, Hupcey, Williams, Bingham. Wiley. ISBN 0470638036
Also, see our posters, talks, and preprints on Nature Precedings: http://precedings.nature.com/account/submissions
