Kara M Dismuke Week 9 Journal
From OpenWetWare
Jump to navigationJump to search
Chapter 4 of Campbell & Heyer (2003): Questions and Answers
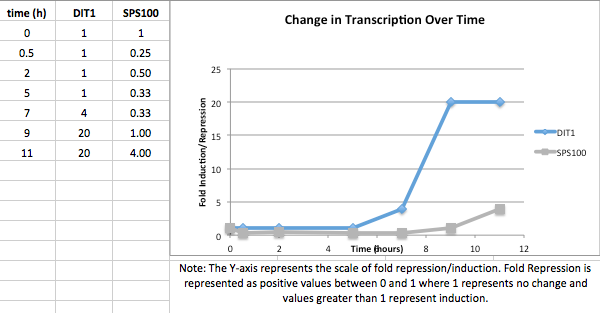
- (Question 5, p. 110) Choose two genes from Figure 4.6b (PDF of figures on MyLMUConnect) and draw a graph to represent the change in transcription over time. You can either create your plot in Excel and put the image up on your wiki page or you can do it in hard copy and turn it in in class.
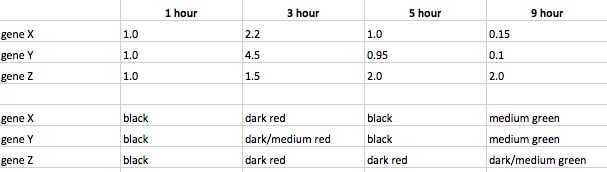
- (Question 6b, p. 110) Look at Figure 4.7, which depicts the loss of oxygen over time and the transcriptional response of three genes. These data are the ratios of transcription for genes X, Y, and Z during the depletion of oxygen. Using the color scale from Figure 4.6, determine the color for each ratio in Figure 4.7b. (Use the nomenclature "bright green", "medium green", "dim green", "black", "dim red", "medium red", or "bright red" for your answers.)
- (Question 7, p. 110) Were any of the genes in Figure 4.7b transcribed similarly? If so, which ones were transcribed similarly to which ones?
- Gene X and gene Y were transcribed similarly as they both started with a ratio of 1.0 at the 1 hour mark. The ratios of both then had increased at the 3 hour mark only to then have decreased at the 5 hour mark back to just about the 1 hour mark ratio levels. By the 9 hour mark, the ratios of both genes were close to 0.
- (Question 9, p. 118) Why would most spots be yellow at the first time point? I.e., what is the technical reason that spots show up as yellow - where does the yellow color come from? And, what would be the biological reason that the experiment resulted in most spots being yellow?
- The yellow spots are a visual way of depicting a red to green ratio of 1:1 (noting yellow is a mix of red and green). Technically, this occurs because there is an equal amount of the control gene and experimental gene present. This occurs initially because the control data is still largely being expressed for not a lot of time has passed. If yellow spots appear at the end of the experiment, then we acknowledge there exists an equal amount of both genes present.
- (Question 10, p. 118) Go to the Saccharomyces Genome Database and search for the gene TEF4; you will see it is involved in translation. Look at the time point labeled OD 3.7 in Figure 4.12, and find the TEF4 spot. Over the course of this experiment, was TEF4 induced or repressed? Hypothesize why TEF4’s change in expression was part of the cell’s response to a reduction in available glucose (i.e., the only available food).
- It appears as though TEF4 was repressed as it changes from yellow to green over the course of the experiment. The TEF4 initiates the metabolism of glucose; however, since there is less glucose present, the system shifts way from this.
- (Question, 11, p. 120) Why would TCA cycle genes be induced if the glucose supply is running out?
- As the glucose supple runs out, it make sense that the TCA cycle genes would be induced as the cell realizes they are approaching starvation, and so. works to conserve the amount of glucose it has (i.e. works to limit the amount of glucose that is broken down).
- (Question 12, p. 120) What mechanism could the genome use to ensure genes for enzymes in a common pathway are induced or repressed simultaneously?
- Clustering allows for organizing all the genes together for further analysis. Upon this procedure, we see that the genes within the cluster may very well have the same repressor or activator in terms of transcription, which would respond to the conditions the cell are in to regulate the production/expression of certain genes.
- (Question 13, p. 121) Consider a microarray experiment where cells deleted for the repressor TUP1 were subjected to the same experiment of a timecourse of glucose depletion where cells at t0 (plenty of glucose available) are labeled green and cells at later timepoints (glucose depleted) are labeled red. What color would you expect the spots that represented glucose-repressed genes to be in the later time points of this experiment?
- Deletion of the repressor TUP1 in certain cells over the course of an experiment where the cell undergoes glucose depletion would, I believe, cause the spots that represented glucose-repressed genes to be red. Since TUP1 suppresses glucose repression, it makes sense that upon its deletion, these genes would exhibit greater expression.
- (Question 14, p. 121) Consider a microarray experiment where cells that overexpress the transcription factor Yap1p were subjected to the same experiment of a timecourse of glucose depletion where cells at t0 (plenty of glucose available) are labeled green and cells at later timepoints (glucose depleted) are labeled red. What color would you expect the spots that represented Yap1p target genes to be in the later time points of this experiment?
- Since YAP1p acts to respond to external stress and the amount of external stress decreases (with the gradual depletion of glucose), more YAP1 is present, and thus, it would expressed as red (it’s particular shade of red would depend on the amount present).
- (Question 16, p. 121) Using the microarray data, how could you verify that you had truly deleted TUP1 or overexpressed YAP1 in the experiments described in questions 8 and 9?
- If TUP1 is truly deleted, then black should appear to indicate the lack of a presence of the gene. If YAP1 is overexpressed, then this would result in bright red as the fold induction would be >20.
Template:Kara M Dismuke
Back to User: User: Kara M Dismuke
- Week 1
- Week 2
- Week 3
- Week 4
- Week 5
- Week 6
- Week 7
- Week 8
- Week 9
- Week 10
- Week 11
- Week 12
- Week 13
- Week 14
- Week 15
- Helpful Pages