JT Correy Journal Week 6
Purpose
The purpose of this assignment was to develop our skills as researchers and progress our understanding of bioinformatics through analyzing and understanding a complex biological system. The binding ability of the SARS-COV-2 to the ACE2 proteins of various species is a complex process and to comprehend it we have to use several techniques we have been introduced to in the class (like modeling and creating of a phylogenetic tree).
Methods
Background research
Background research was done by reading the following articles:
- Daly, N. (2020, April 22). Seven more big cats test positive for coronavirus at Bronx Zoo. Retrieved October 15, 2020, from https://www.nationalgeographic.com/animals/2020/04/tiger-coronavirus-covid19-positive-test-bronx-zoo/
- Zhang, T., Wu, Q., & Zhang, Z. (2020). Probable Pangolin Origin of SARS-CoV-2 Associated with the COVID-19 Outbreak. Current Biology, 30(7). doi:10.1016/j.cub.2020.03.022 from https://www.sciencedirect.com/science/article/pii/S0960982220303602
- Kim, A. (2020, October 13). More than 1 million mink will be killed to help contain a series of Covid-19 outbreaks on Danish farms. Retrieved October 15, 2020, from https://www.cnn.com/2020/10/13/world/denmark-mink-farms-covid-trnd/index.html
- Hoffmann, M., Kleine-Weber, H., Schroeder, S., Krüger, N., Herrler, T., Erichsen, S., . . . Pöhlmann, S. (2020). SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell, 181(2). doi:10.1016/j.cell.2020.02.052 from https://www.sciencedirect.com/science/article/pii/S0092867420302294
- Rees, V. (2020, May 06). Activation sequence of COVID-19 S protein cleaved by furin protease. Retrieved October 15, 2020, from https://www.drugtargetreview.com/news/61264/study-finds-activation-sequence-of-covid-19-s-protein-is-cleaved-by-furin-protease/
- John's Hopkins University, C. (2020, October 15). COVID-19 Map. Retrieved October 15, 2020, from https://coronavirus.jhu.edu/map.html
Sequences
Based on the background research we selected to analyze the following species:
- Human: NP_001358344.1 Homo sapiens
- Rats: AAW78017.1 Rattus norvegicus
- Civet: AAX63775.1 [Paguma larvata
- Mink: U6DXQ3-1 Neovison vison
- Ferret: BAE53380.1 Mustela putorius furo
- Bat: AGZ48803.1 Rhinolophus sinicus
- Mouse: NP_001123985.1 Mus musculus
- Tiger: XP_007090142.1 Panthera tigris altaica
- Pangolin:QLH93383.1 Manis pentadactyla
- Cat: NP_001034545.1 Felis catus
- Dog: NP_001158732.1 Canis lupus familiaris
- Monkey: AAY57872.1 Chlorocebus aethiops
- Pig: NP_001116542.1 Sus scrofa
- Orangutan: Q5RFN1.1 Pongo abelii
We started by searching the for the chosen animal sequences on Unipro
- We had to start using Genbank also because some of the sequences couldn't be found on Unipro
- We searched for "ACE2" and then were able to filter by taxa to go to mammals then to each of our chosen species. We also were able to find some by searching "ACE2 Animal" with the "animal" just being the species we were searching for.
- We had to start using Genbank also because some of the sequences couldn't be found on Unipro
- All the sequences were the respective ACE2 protein sequences. The were downloaded in the FASTA file format.
- The FASTA sequences were then compiled on to one document.
- The sequences were then analyzed to make a phylogenetic tree using Phylogeny.fr
These were the selected mammals and their respective sequences
FASTA format
>NP_001358344.1 [Homo sapiens] MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLASWNYNTNITEENVQNMNNAGDKWS AFLKEQSTLAQMYPLQEIQNLTVKLQLQALQQNGSSVLSEDKSKRLNTILNTMSTIYSTGKVCNPDNPQE CLLLEPGLNEIMANSLDYNERLWAWESWRSEVGKQLRPLYEEYVVLKNEMARANHYEDYGDYWRGDYEVN GVDGYDYSRGQLIEDVEHTFEEIKPLYEHLHAYVRAKLMNAYPSYISPIGCLPAHLLGDMWGRFWTNLYS LTVPFGQKPNIDVTDAMVDQAWDAQRIFKEAEKFFVSVGLPNMTQGFWENSMLTDPGNVQKAVCHPTAWD LGKGDFRILMCTKVTMDDFLTAHHEMGHIQYDMAYAAQPFLLRNGANEGFHEAVGEIMSLSAATPKHLKS IGLLSPDFQEDNETEINFLLKQALTIVGTLPFTYMLEKWRWMVFKGEIPKDQWMKKWWEMKREIVGVVEP VPHDETYCDPASLFHVSNDYSFIRYYTRTLYQFQFQEALCQAAKHEGPLHKCDISNSTEAGQKLFNMLRL GKSEPWTLALENVVGAKNMNVRPLLNYFEPLFTWLKDQNKNSFVGWSTDWSPYADQSIKVRISLKSALGD KAYEWNDNEMYLFRSSVAYAMRQYFLKVKNQMILFGEEDVRVANLKPRISFNFFVTAPKNVSDIIPRTEV EKAIRMSRSRINDAFRLNDNSLEFLGIQPTLGPPNQPPVSIWLIVFGVVMGVIVVGIVILIFTGIRDRKK KNKARSGENPYASIDISKGENNPGFQNTDDVQTSF
>AAW78017.1 [Rattus norvegicus] MSSSCWLLLSLVAVATAQSLIEEKAESFLNKFNQEAEDLSYQSSLASWNYNTNITEENAQKMNEAAAKWS AFYEEQSKIAQNFSLQEIQNATIKRQLKALQQSGSSALSPDKNKQLNTILNTMSTIYSTGKVCNSMNPQE CFLLEPGLDEIMATSTDYNRRLWAWEGWRAEVGKQLRPLYEEYVVLKNEMARANNYEDYGDYWRGDYEAE GVEGYNYNRNQLIEDVENTFKEIKPLYEQLHAYVRTKLMEVYPSYISPTGCLPAHLLGDMWGRFWTNLYP LTTPFLQKPNIDVTDAMVNQSWDAERIFKEAEKFFVSVGLPQMTPGFWTNSMLTEPGDDRKVVCHPTAWD LGHGDFRIKMCTKVTMDNFLTAHHEMGHIQYDMAYAKQPFLLRNGANEGFHEAVGEIMSLSAATPKHLKS IGLLPSNFQEDNETEINFLLKQALTIVGTLPFTYMLEKWRWMVFQDKIPREQWTKKWWEMKREIVGVVEP LPHDETYCDPASLFHVSNDYSFIRYYTRTIYQFQFQEALCQAAKHDGPLHKCDISNSTEAGQKLLNMLSL GNSGPWTLALENVVGSRNMDVKPLLNYFQPLFVWLKEQNRNSTVGWSTDWSPYADQSIKVRISLKSALGK NAYEWTDNEMYLFRSSVAYAMREYFSREKNQTVPFGEADVWVSDLKPRVSFNFFVTSPKNVSDIIPRSEV EEAIRMSRGRINDIFGLNDNSLEFLGIYPTLKPPYEPPVTIWLIIFGVVMGTVVVGIVILIVTGIKGRKK KNETKREENPYDSMDIGKGESNAGFQNSDDAQTSF
>AAX63775.1 [Paguma larvata] MSGSFWLLLSFAALTAAQSTTEELAKTFLETFNYEAQELSYQSSVASWNYNTNITDENAKNMNEAGAKWS AYYEEQSKLAQTYPLAEIQDAKIKRQLQALQQSGSSVLSADKSQRLNTILNAMSTIYSTGKACNPNNPQE CLLLEPGLDNIMENSKDYNERLWAWEGWRAEVGKQLRPLYEEYVALKNEMARANNYEDYGDYWRGDYEEE WTGGYNYSRNQLIQDVEDTFEQIKPLYQHLHAYVRAKLMDTYPSRISRTGCLPAHLLGDMWGRFWTNLYP LTVPFGQKPNIDVTDAMVNQNWDARRIFKEAEKFFVSVGLPNMTQGFWENSMLTEPGDGRKVVCHPTAWD LGKGDFRIKMCTKVTMDDFLTAHHEMGHIQYDMAYAAQPFLLRNGANEGFHEAVGEIMSLSAATPNHLKT IGLLSPAFSEDNETEINFLLKQALTIVGTLPFTYMLEKWRWMVFKGAIPKEQWMQKWWEMKRNIVGVVEP VPHDETYCDPASLFHVANDYSFIRYYTRTIYQFQFQEALCQIAKHEGPLHKCDISNSTEAGKKLLEMLSL GRSEPWTLALERVVGAKNMNVTPLLNYFEPLFTWLKEQNRNSFVGWDTDWRPYSDQSIKVRISLKSALGE KAYEWNDNEMYLFRSSIAYAMREYFSKVKNQTIPFVEDNVWVSDLKPRISFNFFVTFSNNVSDVIPRSEV EDAIRMSRSRINDAFRLDDNSLEFLGIEPTLSPPYRPPVTIWLIVFGVVMGAIVVGIVLLIVSGIRNRRK NDQAGSEENPYASVDLNKGENNPGFQHADDVQTSF
>U6DXQ3-1 [Neovison vison] GLPNMTEGFWQNSMLTEPGDNRKVVCHPTAWDLGKHDFRIKMCTKVTMDDFLTAHHEMGH IQYDMAYAAQPFLLRNGANEGFHEAVGEIMSLSAATPNHLKNIGLLPPDFSEDSETDINF LLKQALTIVGTLPFTYMLEKWRWMVFKGEIPKEQWMQKWWEMKRDIVGVVEPLPHDETYC DPAALFHVANDYSFIRYYTRTIYQFQFQEALCQIAKHEGPLYKCDISNSREAGQKLHEML SLGRSKPWTFALERVVGAKTMDVRPLLNYFEPLFTWLKEQNRNSFVGWNTDWSPYADQSI KVRISLKSALGEKAYEWNDNEMYFFQSSIAYAMREYFSKVKKQTIPFVDKDVRVSDLKPR ISFNFIVTSPENMSDIIPRADVEEAIRKSRGRINDAFRLDDNSLEFLGIQPTLEPPYQPP VTIWLIVFGVVMGVVVVGIFLLIFSGIRNRRKNNQARSEENPYASVDLSKG
>BAE53380.1 [Mustela putorius furo] MLGSSWLLLSLAALTAAQSTTEDLAKTFLEKFNYEAEELSYQNSLASWNYNTNITDENIQKMNIAGAKWS AFYEEESQHAKTYPLEEIQDPIIKRQLRALQQSGSSVLSADKRERLNTILNAMSTIYSTGKACNPNNPQE CLLLEPGLDDIMENSKDYNERLWAWEGWRSEVGKQLRPLYEEYVALKNEMARANNYEDYGDYWRGDYEEE WADGYSYSRNQLIEDVEHTFTQIKPLYEHLHAYVRAKLMDAYPSRISPTGCLPAHLLGDMWGRFWTNLYP LMVPFRQKPNIDVTDAMVNQSWDARRIFEEAETFFVSVGLPNMTEGFWQNSMLTEPGDNRKVVCHPTAWD LGKRDFRIKMCTKVTMDDFLTAHHEMGHIQYDMAYAEQPFLLRNGANEGFHEAVGEIMSLSAATPNHLKN IGLLPPDFSEDSETDINFLLKQALTIVGTLPFTYMLEKWRWMVFKGEIPKEQWMQKWWEMKRDIVGVVEP LPHDETYCDPAALFHVANDYSFIRYYTRTIYQFQFQEALCQIAKHEGPLYKCDISNSSEAGQKLHEMLSL GRSKPWTFALERVVGAKTMDVRPLLNYFEPLFTWLKEQNRNSFVGWNTDWSPYADQSIKVRISLKSALGE KAYEWNDNEMYFFQSSIAYAMREYFSKVKNQTIPFVGKDVRVSDLKPRISFNFIVTSPENMSDIIPRADV EEAIRKSRGRINDAFRLDDNSLEFLGIQPTLEPPYQPPVTIWLIVFGVVMGVVVVGIFLLIFSGIRNRRK NNQARSEENPYASVDLSKGENNPGFQNVDDVQTSF
>AGZ48803.1 [Rhinolophus sinicus] MSGSSWLLLSLVAVTTAQSTTEDEAKMFLDKFNTKAEDLSHQSSLASWDYNTNINDENVQKMDEAGAKWS AFYEEQSKLAKNYSLEQIQNVTVKLQLQILQQSGSPVLSEDKSKRLNSILNAMSTIYSTGKVCKPNKPQE CLLLEPGLDNIMGTSKDYNERLWAWEGWRAEVGKQLRPLYEEYVVLKNEMARGYHYEDYGDYWRRDYETE ESPGPGYSRDQLMKDVERIFTEIKPLYEHLHAYVRAKLMDTYPFHISPTGCLPAHLLGDMWGRFWTNLYP LTVPFGQKPNIDVTDEMLKQGWDADRIFKEAEKFFVSVGLPNMTEGFWNNSMLTEPGDGRKVVCHPTAWD LGKGDFRIKMCTKVTMEDFLTAHHEMGHIQYDMAYASQPYLLRNGANEGFHEAVGEVMSLSVATPKHLKT MGLLSPDFREDNETEINFLLKQALNIVGTLPFTYMLEKWRWMVFKGEIPKEEWMKKWWEMKRKIVGVVEP VPHDETYCDPASLFHVANDYSFIRYYTRTIFEFQFHEALCRIAQHDGPLHKCDISNSTDAGKKLHQMLSV GKSQAWTKTLEDIVDSRNMDVGPLLKYFEPLYTWLQEQNRKSYVGWNTDWSPYSDQSIKVRISLKSALGE NAYEWNDNEMYLFRSSVAYAMREYFLKEKHQTILFGAENVWVSNLKPRISFNFHVTSPGNLSDIIPRPEV EGAIRMSRSRINDAFRLDDNSLEFLGIQPTLGPPYQPPVTIWLIVFGVVMAVVVVGIVVLIITGIRDRRK TDQARSEENPYSSVDLSKGENNPGFQNGDDVQTSF
>NP_001123985.1 [Mus musculus] MSSSSWLLLSLVAVTTAQSLTEENAKTFLNNFNQEAEDLSYQSSLASWNYNTNITEENAQKMSEAAAKWS AFYEEQSKTAQSFSLQEIQTPIIKRQLQALQQSGSSALSADKNKQLNTILNTMSTIYSTGKVCNPKNPQE CLLLEPGLDEIMATSTDYNSRLWAWEGWRAEVGKQLRPLYEEYVVLKNEMARANNYNDYGDYWRGDYEAE GADGYNYNRNQLIEDVERTFAEIKPLYEHLHAYVRRKLMDTYPSYISPTGCLPAHLLGDMWGRFWTNLYP LTVPFAQKPNIDVTDAMMNQGWDAERIFQEAEKFFVSVGLPHMTQGFWANSMLTEPADGRKVVCHPTAWD LGHGDFRIKMCTKVTMDNFLTAHHEMGHIQYDMAYARQPFLLRNGANEGFHEAVGEIMSLSAATPKHLKS IGLLPSDFQEDSETEINFLLKQALTIVGTLPFTYMLEKWRWMVFRGEIPKEQWMKKWWEMKREIVGVVEP LPHDETYCDPASLFHVSNDYSFIRYYTRTIYQFQFQEALCQAAKYNGSLHKCDISNSTEAGQKLLKMLSL GNSEPWTKALENVVGARNMDVKPLLNYFQPLFDWLKEQNRNSFVGWNTEWSPYADQSIKVRISLKSALGA NAYEWTNNEMFLFRSSVAYAMRKYFSIIKNQTVPFLEEDVRVSDLKPRVSFYFFVTSPQNVSDVIPRSEV EDAIRMSRGRINDVFGLNDNSLEFLGIHPTLEPPYQPPVTIWLIIFGVVMALVVVGIIILIVTGIKGRKK KNETKREENPYDSMDIGKGESNAGFQNSDDAQTSF
>XP_007090142.1 [Panthera tigris altaica] LSFAALTAAQSTTEELAKTFLEKFNHEAEELSYQSSLASWNYNTNITDENVQKMNEAGAKWSAFYEEQSK LAETYPLAEIHNTTVKRQLQALQQSGSSVLSADKSQRLNTILNAMSTIYSTGKACNPNNPQECLLLEPGL DDIMENSKDYNERLWAWEGWRAEVGKQLRPLYEEYVALKNEMARANNYEDYGDYWRGDYEEEWTDGYNYS RSQLIKDVEHTFTQIKPLYQHLHAYVRAKLMDSYPSRISPTGCLPAHLLGDMWGRFWTNLYPLTVPFGQK PNIDVTDAMVNQSWDARRIFKEAEKFFVSVGLPNMTQGFWENSMLTEPGNSQKVVCHPTAWDLGKGDFRI KMCTKVTMDDFLTAHHEMGHIQYDMAYAVQPFLLRNGANEGFHEAVGEIMSLSAATPNHLKTIGLLPPGF SEDSETEINFLLKQALTIVGTLPFTYMLEKWRWMVFKGEIPKEQWMQKWWEMKREIVGVVEPVPHDETYC DPASLFHVANDYSFIRYYTRTIYQFQFQEALCRIAKHEGPLHKCDISNSSEAGKKLLQMLTLGKSKPWTL ALEHVVGEKNMNVTPLLKYFEPLFTWLKEQNRNSFVGWNTDWRPYADQSIKVRISLKSALGDKAYEWNDN EMYLFRSSVAYAMREYFSKVKNQTIPFVEDNVWVSNLKPRISFNFFVTASKNVSDVIPRREVEEAIRMSR SRINDAFRLDDNSLEFLGIQPTLSPPYQPPVTIWLIVFGVVMGVVVVGIVLLIVSGIRNRRKNNQARSEE NPYASVDLSKGENNPGFQHADDVQTSF
>QLH93383.1 [Manis pentadactyla] MSGSSWLLLSLVAVTAAQSTSDEEAKTFLEKFNSEAEELSYQSSLASWNYNTNITDENVQKMNVAGAKWS TFYEEQSKIAKNYQLQNIQNDTIKRQLQALQLSGSSALSADKNQRLNTILNTMSTIYSTGKVCNPGNPQE CSLLEPGLDNIMESSKDYNERLWAWEGWRSEVGKQLRPLYEEYVVLKNEMARANHYEDYGDYWRGDYETE GANGYNYSRDHLIEDVEHIFTQIKPLYEHLHAYVRAKLMDNYPSHISPTGCLPAHLLGDMWGRFWTNLYP LTVPFRQKPNIDVTDAMVNQTWDANRIFKEAEKFFVSVGLPKMTQTFWENSMLTEPGDGRKVVCHPTAWD LGKHDFRIKMCTKVTMDDFLTAHHEMGHIQYDMAYAMQPYLLRNGANEGFHEAVGEIMSLSAATPKHLKN IGLLPPDFYEDNETEINFLLKQALTIVGTLPFTYMLEKWRWMVFSGQIPKEQWMKKWWEMKREIVGVVEP VPHDETYCDPASLFHVANDYSFIRYYTRTIYQFQFQEALCQTAKHEGPLHKCDISNSTEAGQKLLQMLSL GKSKPWTLALERVVGTKNMDVRPLLNYFEPLLTWLKEQNKNSFVGWNTDWSPYAAQSIKVRISLKSALGE KAYEWNDSEMYLFRSSVAYAMREYFSKFKKQTIPFEEESVRVSDLKPRVSFIFFVTLPKNVSAVIPRAEV EEAIRMSRSRINDVFRLDDNSLEFLGIQPTLEPPYQPPVTIWLIVFGVVMGVIVVGIVVLIFTGIRDRKK KNQARSEQNPYASVDLSKGENNPGFQNVDDVQTSF
>NP_001034545.1 [Felis catus] MSGSFWLLLSFAALTAAQSTTEELAKTFLEKFNHEAEELSYQSSLASWNYNTNITDENVQKMNEAGAKWS AFYEEQSKLAKTYPLAEIHNTTVKRQLQALQQSGSSVLSADKSQRLNTILNAMSTIYSTGKACNPNNPQE CLLLEPGLDDIMENSKDYNERLWAWEGWRAEVGKQLRPLYEEYVALKNEMARANNYEDYGDYWRGDYEEE WTDGYNYSRSQLIKDVEHTFTQIKPLYQHLHAYVRAKLMDTYPSRISPTGCLPAHLLGDMWGRFWTNLYP LTVPFGQKPNIDVTDAMVNQSWDARRIFKEAEKFFVSVGLPNMTQGFWENSMLTEPGDSRKVVCHPTAWD LGKGDFRIKMCTKVTMDDFLTAHHEMGHIQYDMAYAVQPFLLRNGANEGFHEAVGEIMSLSAATPNHLKT IGLLSPGFSEDSETEINFLLKQALTIVGTLPFTYMLEKWRWMVFKGEIPKEQWMQKWWEMKREIVGVVEP VPHDETYCDPASLFHVANDYSFIRYYTRTIYQFQFQEALCRIAKHEGPLHKCDISNSSEAGKKLLQMLTL GKSKPWTLALEHVVGEKKMNVTPLLKYFEPLFTWLKEQNRNSFVGWNTDWRPYADQSIKVRISLKSALGD EAYEWNDNEMYLFRSSVAYAMREYFSKVKNQTIPFVEDNVWVSNLKPRISFNFFVTASKNVSDVIPRSEV EEAIRMSRSRINDAFRLDDNSLEFLGIQPTLSPPYQPPVTIWLIVFGVVMGVVVVGIVLLIVSGIRNRRK NNQARSEENPYASVDLSKGENNPGFQHADDVQTSF
>NP_001158732.1 [Canis lupus familiaris] MSGSSWLLLSLAALTAAQSTEDLVKTFLEKFNYEAEELSYQSSLASWNYNINITDENVQKMNNAGAKWSA FYEEQSKLAKTYPLEEIQDSTVKRQLRALQHSGSSVLSADKNQRLNTILNSMSTVYSTGKACNPSNPQEC LLLEPGLDDIMENSKDYNERLWAWEGWRSEVGKQLRPLYEEYVALKNEMARANNYEDYGDYWRGDYEEEW ENGYNYSRNQLIDDVELTFTQIMPLYQHLHAYVRTKLMDTYPSYISPTGCLPAHLLGDMWGRFWTNLYPL TVPFGQKPNIDVTNAMVNQSWDARKIFKEAEKFFVSVGLPNMTQEFWGNSMLTEPSDSRKVVCHPTAWDL GKGDFRIKMCTKVTMDDFLTAHHEMGHIQYDMAYAAQPFLLRNGANEGFHEAVGEIMSLSAATPNHLKNI GLLPPSFFEDSETEINFLLKQALTIVGTLPFTYMLEKWRWMVFKGEIPKDQWMKTWWEMKRNIVGVVEPV PHDETYCDPASLFHVANDYSFIRYYTRTIYQFQFQEALCQIAKHEGPLHKCDISNSSEAGQKLLEMLKLG KSKPWTYALEIVVGAKNMDVRPLLNYFEPLFTWLKEQNRNSFVGWNTDWSPYADQSIKVRISLKSALGEK AYEWNNNEMYLFRSSIAYAMRQYFSEVKNQTIPFVEDNVWVSDLKPRISFNFSVTSPGNVSDIIPRTEVE EAIRMYRSRINDVFRLDDNSLEFLGIQPTPGPPYEPPVTIWLIVFGVVMGVVVVGIVLLIFSGIRNRRKN DQARGEENPYASVDLSKGENNPGFQSGDDVQTSF
>AAY57872.1 [Chlorocebus aethiops] MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLASWNYNTNITEENVQNMNNAGEKWS AFLKEQSTLAQMYPLQAIQNLTVKLQLQALQQNGSSVLSEDKSKRLNTILNTMSTIHSTGKVCNPNNPQE CLLLDPGLNEIMEKSLDYNERLWAWEGWRSEVGKQLRPLYEEYVVLKNEMARANHYKDYGDYWRGDYEVN GVDGYDYNRDQLIEDVERTFEEIKPLYEHLHAYVRAKLMNAYPSYISPTGCLPAHLLGDMWGRFWTNLYS LTVPFGQKPNIDVTDAMVNQAWNAQRIFKEAEKFFVSVGLPNMTQGFWENSMLTDPGNVQKVVCHPTAWD LGKGDFRIIMCTKVTMDDFLTAHHEMGHIQYDMAYAAQPFLLRNGANEGFHEAVGEIMSLSAATPKHLKS IGLLSPDFQEDNETEINFLLKQALTIVGTLPFTYMLEKWRWMVFKGEIPKDQWMKKWWEMKREIVGVVEP VPHDETYCDPASLFHVSNDYSFIRYYTRTLYQFQFQEALCQAAKHEGPLHKCDISNSTEAGQKLLNMLKL GKSEPWTLALENVVGAKNMSVRPLLNYFEPLFTWLKDQNKNSFVGWSTDWSPYADQSIKVRISLKSALGA NAYKWNDNEMYLFRSSVAYAMRQYFLENKHQTILFGEEDVRVADLKPRISFNFYVTAPKNVSDIIPRTEV EEAIRFSRSRINDAFQLNDNSLEFLGIQSTLVPPYQSPITTWLIVFGVVMAVIVAGIVVLIFTGIRDRKK KNQARSEENPYASIDISKGENNPGFQNTDDVQTSF
>NP_001116542.1 [Sus scrofa] MSGSFWLLLSLIPVTAAQSTTEELAKTFLEKFNLEAEDLAYQSSLASWTINTNITDENIQKMNDARAKWS AFYEEQSRIAKTYPLDEIQTLILKRQLQALQQSGTSGLSADKSKRLNTILNTMSTIYSSGKVLDPNNPQE CLVLEPGLDEIMENSKDYSRRLWAWESWRAEVGKQLRPLYEEYVVLENEMARANNYEDYGDYWRGDYEVT GTGDYDYSRNQLMEDVERTFAEIKPLYEHLHAYVRAKLMDAYPSRISPTGCLPAHLLGDMWGRFWTNLYP LTVPFGEKPSIDVTEAMVNQSWDAIRIFEEAEKFFVSIGLPNMTQGFWNNSMLTEPGDGRKVVCHPTAWD LGKGDFRIKMCTKVTMDDFLTAHHEMGHIQYDMAYAIQPYLLRNGANEGFHEAVGEIMSLSAATPHYLKA LGLLPPDFYEDSETEINFLLKQALTIVGTLPFTYMLEKWRWMVFKGEIPKEQWMQKWWEMKREIVGVVEP LPHDETYCDPACLFHVAEDYSFIRYYTRTIYQFQFHEALCRTAKHEGPLYKCDISNSTEAGQKLLQMLSL GKSEPWTLALENIVGVKTMDVKPLLSYFEPLLTWLKAQNGNSSVGWNTDWTPYADQSIKVRISLKSALGE DAYEWNDNEMYLFRSSIAYAMRNYFSSAKNETIPFGAVDVWVSDLKPRISFNFFVTSPANMSDIIPRSDV EKAISMSRSRINDAFRLDDNTLEFLGIQPTLGPPDEPPVTVWLIIFGVVMGLVVVGIVVLIFTGIRDRRK KKQASSEENPYGSMDLSKGESNSGFQNGDDIQTSF
>Q5RFN1.1 [Pongo abelii] MSGSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLASWNYNTNITEENVQNMNNAGDKWS AFLKEQSTLAQMYPLQEIQNLTVKLQLQALQQNGSSVLSEDKSKRLNTILNTMSTIYSTGKVCNPNNPQE CLLLEPGLNEIMANSLDYNERLWAWESWRSEVGKQLRPLYEEYVVLKNEMARANHYEDYGDYWRGDYEVN GVDSYDYSRGQLIEDVEHTFEEIKPLYEHLHAYVRAKLINAYPSYISPIGCLPAHLLGDMWGRFWTNLYS LTVPFGQKPNIDVTDAMVDQAWDAQRIFKEAEKFFVSVGLPNMTQRFWENSMLTDPGNVQKVVCHPTAWD LGKGDFRILMCTKVTMDDFLTAHHEMGHIQYDMAYAAQPFLLRNGANEGFHEAVGEIMSLSAATPKHLKS IGLLSPDFQEDNETEINFLLKQALTIVGTLPFTYMLEKWRWMVFKGEIPKDQWMKKWWEMKREIVGVVEP VPHDETYCDPASLFHVSNDYSFIRYYTRTLYQFQFQEALCQAAKHEGPLHKCDISNSTEAGQKLLNMLRL GKSEPWTLALENVVGAKNMNVRPLLDYFEPLFTWLKDQNKNSFVGWSTDWSPYADQSIKVRISLKSALGN KAYEWNDNEIYLFRSSVAYAMRKYFLEVKNQMILFGEEDVRVANLKPRISFNFFVTAPKNVSDIIPRTEV EKAIRMSRSRINDAFRLNDNSLEFLGIQPTLGPPNQPPVSIWLIVFGVVMGVIVVGIVVLIFTGIRDRKK KNKARNEENPYASIDISKGENNPGFQNTDDVQTSF
Clustal Format
The sequences were the aligned into the clustal format for better viewing. This was done with Phylogeny.fr.
AAW78017.1 MSSSCWLLLSLVAVATAQSLIEEKAESFLNKFNQEAEDLSYQSSLASWNYNTNITEENAQ
NP_0011239 MSSSSWLLLSLVAVTTAQSLTEENAKTFLNNFNQEAEDLSYQSSLASWNYNTNITEENAQ
AGZ48803.1 MSGSSWLLLSLVAVTTAQSTTEDEAKMFLDKFNTKAEDLSHQSSLASWDYNTNINDENVQ
NP_0011165 MSGSFWLLLSLIPVTAAQSTTEELAKTFLEKFNLEAEDLAYQSSLASWTINTNITDENIQ
AAY57872.1 MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLASWNYNTNITEENVQ
NP_0013583 MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLASWNYNTNITEENVQ
Q5RFN1.1_P MSGSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLASWNYNTNITEENVQ
QLH93383.1 MSGSSWLLLSLVAVTAAQSTSDEEAKTFLEKFNSEAEELSYQSSLASWNYNTNITDENVQ
U6DXQ3-1_N ------------------------------------------------------------
BAE53380.1 MLGSSWLLLSLAALTAAQSTTEDLAKTFLEKFNYEAEELSYQNSLASWNYNTNITDENIQ
NP_0011587 MSGSSWLLLSLAALTAAQST-EDLVKTFLEKFNYEAEELSYQSSLASWNYNINITDENVQ
AAX63775.1 MSGSFWLLLSFAALTAAQSTTEELAKTFLETFNYEAQELSYQSSVASWNYNTNITDENAK
XP_0070901 --------LSFAALTAAQSTTEELAKTFLEKFNHEAEELSYQSSLASWNYNTNITDENVQ
NP_0010345 MSGSFWLLLSFAALTAAQSTTEELAKTFLEKFNHEAEELSYQSSLASWNYNTNITDENVQ
AAW78017.1 KMNEAAAKWSAFYEEQSKIAQNFSLQEIQNATIKRQLKALQQSGSSALSPDKNKQLNTIL
NP_0011239 KMSEAAAKWSAFYEEQSKTAQSFSLQEIQTPIIKRQLQALQQSGSSALSADKNKQLNTIL
AGZ48803.1 KMDEAGAKWSAFYEEQSKLAKNYSLEQIQNVTVKLQLQILQQSGSPVLSEDKSKRLNSIL
NP_0011165 KMNDARAKWSAFYEEQSRIAKTYPLDEIQTLILKRQLQALQQSGTSGLSADKSKRLNTIL
AAY57872.1 NMNNAGEKWSAFLKEQSTLAQMYPLQAIQNLTVKLQLQALQQNGSSVLSEDKSKRLNTIL
NP_0013583 NMNNAGDKWSAFLKEQSTLAQMYPLQEIQNLTVKLQLQALQQNGSSVLSEDKSKRLNTIL
Q5RFN1.1_P NMNNAGDKWSAFLKEQSTLAQMYPLQEIQNLTVKLQLQALQQNGSSVLSEDKSKRLNTIL
QLH93383.1 KMNVAGAKWSTFYEEQSKIAKNYQLQNIQNDTIKRQLQALQLSGSSALSADKNQRLNTIL
U6DXQ3-1_N ------------------------------------------------------------
BAE53380.1 KMNIAGAKWSAFYEEESQHAKTYPLEEIQDPIIKRQLRALQQSGSSVLSADKRERLNTIL
NP_0011587 KMNNAGAKWSAFYEEQSKLAKTYPLEEIQDSTVKRQLRALQHSGSSVLSADKNQRLNTIL
AAX63775.1 NMNEAGAKWSAYYEEQSKLAQTYPLAEIQDAKIKRQLQALQQSGSSVLSADKSQRLNTIL
XP_0070901 KMNEAGAKWSAFYEEQSKLAETYPLAEIHNTTVKRQLQALQQSGSSVLSADKSQRLNTIL
NP_0010345 KMNEAGAKWSAFYEEQSKLAKTYPLAEIHNTTVKRQLQALQQSGSSVLSADKSQRLNTIL
AAW78017.1 NTMSTIYSTGKVCNSMNPQECFLLEPGLDEIMATSTDYNRRLWAWEGWRAEVGKQLRPLY
NP_0011239 NTMSTIYSTGKVCNPKNPQECLLLEPGLDEIMATSTDYNSRLWAWEGWRAEVGKQLRPLY
AGZ48803.1 NAMSTIYSTGKVCKPNKPQECLLLEPGLDNIMGTSKDYNERLWAWEGWRAEVGKQLRPLY
NP_0011165 NTMSTIYSSGKVLDPNNPQECLVLEPGLDEIMENSKDYSRRLWAWESWRAEVGKQLRPLY
AAY57872.1 NTMSTIHSTGKVCNPNNPQECLLLDPGLNEIMEKSLDYNERLWAWEGWRSEVGKQLRPLY
NP_0013583 NTMSTIYSTGKVCNPDNPQECLLLEPGLNEIMANSLDYNERLWAWESWRSEVGKQLRPLY
Q5RFN1.1_P NTMSTIYSTGKVCNPNNPQECLLLEPGLNEIMANSLDYNERLWAWESWRSEVGKQLRPLY
QLH93383.1 NTMSTIYSTGKVCNPGNPQECSLLEPGLDNIMESSKDYNERLWAWEGWRSEVGKQLRPLY
U6DXQ3-1_N ------------------------------------------------------------
BAE53380.1 NAMSTIYSTGKACNPNNPQECLLLEPGLDDIMENSKDYNERLWAWEGWRSEVGKQLRPLY
NP_0011587 NSMSTVYSTGKACNPSNPQECLLLEPGLDDIMENSKDYNERLWAWEGWRSEVGKQLRPLY
AAX63775.1 NAMSTIYSTGKACNPNNPQECLLLEPGLDNIMENSKDYNERLWAWEGWRAEVGKQLRPLY
XP_0070901 NAMSTIYSTGKACNPNNPQECLLLEPGLDDIMENSKDYNERLWAWEGWRAEVGKQLRPLY
NP_0010345 NAMSTIYSTGKACNPNNPQECLLLEPGLDDIMENSKDYNERLWAWEGWRAEVGKQLRPLY
AAW78017.1 EEYVVLKNEMARANNYEDYGDYWRGDYEAEGVEGYNYNRNQLIEDVENTFKEIKPLYEQL
NP_0011239 EEYVVLKNEMARANNYNDYGDYWRGDYEAEGADGYNYNRNQLIEDVERTFAEIKPLYEHL
AGZ48803.1 EEYVVLKNEMARGYHYEDYGDYWRRDYETEESPGPGYSRDQLMKDVERIFTEIKPLYEHL
NP_0011165 EEYVVLENEMARANNYEDYGDYWRGDYEVTGTGDYDYSRNQLMEDVERTFAEIKPLYEHL
AAY57872.1 EEYVVLKNEMARANHYKDYGDYWRGDYEVNGVDGYDYNRDQLIEDVERTFEEIKPLYEHL
NP_0013583 EEYVVLKNEMARANHYEDYGDYWRGDYEVNGVDGYDYSRGQLIEDVEHTFEEIKPLYEHL
Q5RFN1.1_P EEYVVLKNEMARANHYEDYGDYWRGDYEVNGVDSYDYSRGQLIEDVEHTFEEIKPLYEHL
QLH93383.1 EEYVVLKNEMARANHYEDYGDYWRGDYETEGANGYNYSRDHLIEDVEHIFTQIKPLYEHL
U6DXQ3-1_N ------------------------------------------------------------
BAE53380.1 EEYVALKNEMARANNYEDYGDYWRGDYEEEWADGYSYSRNQLIEDVEHTFTQIKPLYEHL
NP_0011587 EEYVALKNEMARANNYEDYGDYWRGDYEEEWENGYNYSRNQLIDDVELTFTQIMPLYQHL
AAX63775.1 EEYVALKNEMARANNYEDYGDYWRGDYEEEWTGGYNYSRNQLIQDVEDTFEQIKPLYQHL
XP_0070901 EEYVALKNEMARANNYEDYGDYWRGDYEEEWTDGYNYSRSQLIKDVEHTFTQIKPLYQHL
NP_0010345 EEYVALKNEMARANNYEDYGDYWRGDYEEEWTDGYNYSRSQLIKDVEHTFTQIKPLYQHL
AAW78017.1 HAYVRTKLMEVYPSYISPTGCLPAHLLGDMWGRFWTNLYPLTTPFLQKPNIDVTDAMVNQ
NP_0011239 HAYVRRKLMDTYPSYISPTGCLPAHLLGDMWGRFWTNLYPLTVPFAQKPNIDVTDAMMNQ
AGZ48803.1 HAYVRAKLMDTYPFHISPTGCLPAHLLGDMWGRFWTNLYPLTVPFGQKPNIDVTDEMLKQ
NP_0011165 HAYVRAKLMDAYPSRISPTGCLPAHLLGDMWGRFWTNLYPLTVPFGEKPSIDVTEAMVNQ
AAY57872.1 HAYVRAKLMNAYPSYISPTGCLPAHLLGDMWGRFWTNLYSLTVPFGQKPNIDVTDAMVNQ
NP_0013583 HAYVRAKLMNAYPSYISPIGCLPAHLLGDMWGRFWTNLYSLTVPFGQKPNIDVTDAMVDQ
Q5RFN1.1_P HAYVRAKLINAYPSYISPIGCLPAHLLGDMWGRFWTNLYSLTVPFGQKPNIDVTDAMVDQ
QLH93383.1 HAYVRAKLMDNYPSHISPTGCLPAHLLGDMWGRFWTNLYPLTVPFRQKPNIDVTDAMVNQ
U6DXQ3-1_N ------------------------------------------------------------
BAE53380.1 HAYVRAKLMDAYPSRISPTGCLPAHLLGDMWGRFWTNLYPLMVPFRQKPNIDVTDAMVNQ
NP_0011587 HAYVRTKLMDTYPSYISPTGCLPAHLLGDMWGRFWTNLYPLTVPFGQKPNIDVTNAMVNQ
AAX63775.1 HAYVRAKLMDTYPSRISRTGCLPAHLLGDMWGRFWTNLYPLTVPFGQKPNIDVTDAMVNQ
XP_0070901 HAYVRAKLMDSYPSRISPTGCLPAHLLGDMWGRFWTNLYPLTVPFGQKPNIDVTDAMVNQ
NP_0010345 HAYVRAKLMDTYPSRISPTGCLPAHLLGDMWGRFWTNLYPLTVPFGQKPNIDVTDAMVNQ
AAW78017.1 SWDAERIFKEAEKFFVSVGLPQMTPGFWTNSMLTEPGDDRKVVCHPTAWDLGHGDFRIKM
NP_0011239 GWDAERIFQEAEKFFVSVGLPHMTQGFWANSMLTEPADGRKVVCHPTAWDLGHGDFRIKM
AGZ48803.1 GWDADRIFKEAEKFFVSVGLPNMTEGFWNNSMLTEPGDGRKVVCHPTAWDLGKGDFRIKM
NP_0011165 SWDAIRIFEEAEKFFVSIGLPNMTQGFWNNSMLTEPGDGRKVVCHPTAWDLGKGDFRIKM
AAY57872.1 AWNAQRIFKEAEKFFVSVGLPNMTQGFWENSMLTDPGNVQKVVCHPTAWDLGKGDFRIIM
NP_0013583 AWDAQRIFKEAEKFFVSVGLPNMTQGFWENSMLTDPGNVQKAVCHPTAWDLGKGDFRILM
Q5RFN1.1_P AWDAQRIFKEAEKFFVSVGLPNMTQRFWENSMLTDPGNVQKVVCHPTAWDLGKGDFRILM
QLH93383.1 TWDANRIFKEAEKFFVSVGLPKMTQTFWENSMLTEPGDGRKVVCHPTAWDLGKHDFRIKM
U6DXQ3-1_N ------------------GLPNMTEGFWQNSMLTEPGDNRKVVCHPTAWDLGKHDFRIKM
BAE53380.1 SWDARRIFEEAETFFVSVGLPNMTEGFWQNSMLTEPGDNRKVVCHPTAWDLGKRDFRIKM
NP_0011587 SWDARKIFKEAEKFFVSVGLPNMTQEFWGNSMLTEPSDSRKVVCHPTAWDLGKGDFRIKM
AAX63775.1 NWDARRIFKEAEKFFVSVGLPNMTQGFWENSMLTEPGDGRKVVCHPTAWDLGKGDFRIKM
XP_0070901 SWDARRIFKEAEKFFVSVGLPNMTQGFWENSMLTEPGNSQKVVCHPTAWDLGKGDFRIKM
NP_0010345 SWDARRIFKEAEKFFVSVGLPNMTQGFWENSMLTEPGDSRKVVCHPTAWDLGKGDFRIKM
***:** ** *****:*.: .*.**********: **** *
AAW78017.1 CTKVTMDNFLTAHHEMGHIQYDMAYAKQPFLLRNGANEGFHEAVGEIMSLSAATPKHLKS
NP_0011239 CTKVTMDNFLTAHHEMGHIQYDMAYARQPFLLRNGANEGFHEAVGEIMSLSAATPKHLKS
AGZ48803.1 CTKVTMEDFLTAHHEMGHIQYDMAYASQPYLLRNGANEGFHEAVGEVMSLSVATPKHLKT
NP_0011165 CTKVTMDDFLTAHHEMGHIQYDMAYAIQPYLLRNGANEGFHEAVGEIMSLSAATPHYLKA
AAY57872.1 CTKVTMDDFLTAHHEMGHIQYDMAYAAQPFLLRNGANEGFHEAVGEIMSLSAATPKHLKS
NP_0013583 CTKVTMDDFLTAHHEMGHIQYDMAYAAQPFLLRNGANEGFHEAVGEIMSLSAATPKHLKS
Q5RFN1.1_P CTKVTMDDFLTAHHEMGHIQYDMAYAAQPFLLRNGANEGFHEAVGEIMSLSAATPKHLKS
QLH93383.1 CTKVTMDDFLTAHHEMGHIQYDMAYAMQPYLLRNGANEGFHEAVGEIMSLSAATPKHLKN
U6DXQ3-1_N CTKVTMDDFLTAHHEMGHIQYDMAYAAQPFLLRNGANEGFHEAVGEIMSLSAATPNHLKN
BAE53380.1 CTKVTMDDFLTAHHEMGHIQYDMAYAEQPFLLRNGANEGFHEAVGEIMSLSAATPNHLKN
NP_0011587 CTKVTMDDFLTAHHEMGHIQYDMAYAAQPFLLRNGANEGFHEAVGEIMSLSAATPNHLKN
AAX63775.1 CTKVTMDDFLTAHHEMGHIQYDMAYAAQPFLLRNGANEGFHEAVGEIMSLSAATPNHLKT
XP_0070901 CTKVTMDDFLTAHHEMGHIQYDMAYAVQPFLLRNGANEGFHEAVGEIMSLSAATPNHLKT
NP_0010345 CTKVTMDDFLTAHHEMGHIQYDMAYAVQPFLLRNGANEGFHEAVGEIMSLSAATPNHLKT
******::****************** **:****************:****.***::**
AAW78017.1 IGLLPSNFQEDNETEINFLLKQALTIVGTLPFTYMLEKWRWMVFQDKIPREQWTKKWWEM
NP_0011239 IGLLPSDFQEDSETEINFLLKQALTIVGTLPFTYMLEKWRWMVFRGEIPKEQWMKKWWEM
AGZ48803.1 MGLLSPDFREDNETEINFLLKQALNIVGTLPFTYMLEKWRWMVFKGEIPKEEWMKKWWEM
NP_0011165 LGLLPPDFYEDSETEINFLLKQALTIVGTLPFTYMLEKWRWMVFKGEIPKEQWMQKWWEM
AAY57872.1 IGLLSPDFQEDNETEINFLLKQALTIVGTLPFTYMLEKWRWMVFKGEIPKDQWMKKWWEM
NP_0013583 IGLLSPDFQEDNETEINFLLKQALTIVGTLPFTYMLEKWRWMVFKGEIPKDQWMKKWWEM
Q5RFN1.1_P IGLLSPDFQEDNETEINFLLKQALTIVGTLPFTYMLEKWRWMVFKGEIPKDQWMKKWWEM
QLH93383.1 IGLLPPDFYEDNETEINFLLKQALTIVGTLPFTYMLEKWRWMVFSGQIPKEQWMKKWWEM
U6DXQ3-1_N IGLLPPDFSEDSETDINFLLKQALTIVGTLPFTYMLEKWRWMVFKGEIPKEQWMQKWWEM
BAE53380.1 IGLLPPDFSEDSETDINFLLKQALTIVGTLPFTYMLEKWRWMVFKGEIPKEQWMQKWWEM
NP_0011587 IGLLPPSFFEDSETEINFLLKQALTIVGTLPFTYMLEKWRWMVFKGEIPKDQWMKTWWEM
AAX63775.1 IGLLSPAFSEDNETEINFLLKQALTIVGTLPFTYMLEKWRWMVFKGAIPKEQWMQKWWEM
XP_0070901 IGLLPPGFSEDSETEINFLLKQALTIVGTLPFTYMLEKWRWMVFKGEIPKEQWMQKWWEM
NP_0010345 IGLLSPGFSEDSETEINFLLKQALTIVGTLPFTYMLEKWRWMVFKGEIPKEQWMQKWWEM
:***.. * **.**:*********.******************* . **.::* :.****
AAW78017.1 KREIVGVVEPLPHDETYCDPASLFHVSNDYSFIRYYTRTIYQFQFQEALCQAAKHDGPLH
NP_0011239 KREIVGVVEPLPHDETYCDPASLFHVSNDYSFIRYYTRTIYQFQFQEALCQAAKYNGSLH
AGZ48803.1 KRKIVGVVEPVPHDETYCDPASLFHVANDYSFIRYYTRTIFEFQFHEALCRIAQHDGPLH
NP_0011165 KREIVGVVEPLPHDETYCDPACLFHVAEDYSFIRYYTRTIYQFQFHEALCRTAKHEGPLY
AAY57872.1 KREIVGVVEPVPHDETYCDPASLFHVSNDYSFIRYYTRTLYQFQFQEALCQAAKHEGPLH
NP_0013583 KREIVGVVEPVPHDETYCDPASLFHVSNDYSFIRYYTRTLYQFQFQEALCQAAKHEGPLH
Q5RFN1.1_P KREIVGVVEPVPHDETYCDPASLFHVSNDYSFIRYYTRTLYQFQFQEALCQAAKHEGPLH
QLH93383.1 KREIVGVVEPVPHDETYCDPASLFHVANDYSFIRYYTRTIYQFQFQEALCQTAKHEGPLH
U6DXQ3-1_N KRDIVGVVEPLPHDETYCDPAALFHVANDYSFIRYYTRTIYQFQFQEALCQIAKHEGPLY
BAE53380.1 KRDIVGVVEPLPHDETYCDPAALFHVANDYSFIRYYTRTIYQFQFQEALCQIAKHEGPLY
NP_0011587 KRNIVGVVEPVPHDETYCDPASLFHVANDYSFIRYYTRTIYQFQFQEALCQIAKHEGPLH
AAX63775.1 KRNIVGVVEPVPHDETYCDPASLFHVANDYSFIRYYTRTIYQFQFQEALCQIAKHEGPLH
XP_0070901 KREIVGVVEPVPHDETYCDPASLFHVANDYSFIRYYTRTIYQFQFQEALCRIAKHEGPLH
NP_0010345 KREIVGVVEPVPHDETYCDPASLFHVANDYSFIRYYTRTIYQFQFQEALCRIAKHEGPLH
**.*******:**********.****::***********:::***:****. *:::*.*:
AAW78017.1 KCDISNSTEAGQKLLNMLSLGNSGPWTLALENVVGSRNMDVKPLLNYFQPLFVWLKEQNR
NP_0011239 KCDISNSTEAGQKLLKMLSLGNSEPWTKALENVVGARNMDVKPLLNYFQPLFDWLKEQNR
AGZ48803.1 KCDISNSTDAGKKLHQMLSVGKSQAWTKTLEDIVDSRNMDVGPLLKYFEPLYTWLQEQNR
NP_0011165 KCDISNSTEAGQKLLQMLSLGKSEPWTLALENIVGVKTMDVKPLLSYFEPLLTWLKAQNG
AAY57872.1 KCDISNSTEAGQKLLNMLKLGKSEPWTLALENVVGAKNMSVRPLLNYFEPLFTWLKDQNK
NP_0013583 KCDISNSTEAGQKLFNMLRLGKSEPWTLALENVVGAKNMNVRPLLNYFEPLFTWLKDQNK
Q5RFN1.1_P KCDISNSTEAGQKLLNMLRLGKSEPWTLALENVVGAKNMNVRPLLDYFEPLFTWLKDQNK
QLH93383.1 KCDISNSTEAGQKLLQMLSLGKSKPWTLALERVVGTKNMDVRPLLNYFEPLLTWLKEQNK
U6DXQ3-1_N KCDISNSREAGQKLHEMLSLGRSKPWTFALERVVGAKTMDVRPLLNYFEPLFTWLKEQNR
BAE53380.1 KCDISNSSEAGQKLHEMLSLGRSKPWTFALERVVGAKTMDVRPLLNYFEPLFTWLKEQNR
NP_0011587 KCDISNSSEAGQKLLEMLKLGKSKPWTYALEIVVGAKNMDVRPLLNYFEPLFTWLKEQNR
AAX63775.1 KCDISNSTEAGKKLLEMLSLGRSEPWTLALERVVGAKNMNVTPLLNYFEPLFTWLKEQNR
XP_0070901 KCDISNSSEAGKKLLQMLTLGKSKPWTLALEHVVGEKNMNVTPLLKYFEPLFTWLKEQNR
NP_0010345 KCDISNSSEAGKKLLQMLTLGKSKPWTLALEHVVGEKKMNVTPLLKYFEPLFTWLKEQNR
******* :**:** :** :*.* .** :** :*. ..*.* ***.**:** **: **
AAW78017.1 NSTVGWSTDWSPYADQSIKVRISLKSALGKNAYEWTDNEMYLFRSSVAYAMREYFSREKN
NP_0011239 NSFVGWNTEWSPYADQSIKVRISLKSALGANAYEWTNNEMFLFRSSVAYAMRKYFSIIKN
AGZ48803.1 KSYVGWNTDWSPYSDQSIKVRISLKSALGENAYEWNDNEMYLFRSSVAYAMREYFLKEKH
NP_0011165 NSSVGWNTDWTPYADQSIKVRISLKSALGEDAYEWNDNEMYLFRSSIAYAMRNYFSSAKN
AAY57872.1 NSFVGWSTDWSPYADQSIKVRISLKSALGANAYKWNDNEMYLFRSSVAYAMRQYFLENKH
NP_0013583 NSFVGWSTDWSPYADQSIKVRISLKSALGDKAYEWNDNEMYLFRSSVAYAMRQYFLKVKN
Q5RFN1.1_P NSFVGWSTDWSPYADQSIKVRISLKSALGNKAYEWNDNEIYLFRSSVAYAMRKYFLEVKN
QLH93383.1 NSFVGWNTDWSPYAAQSIKVRISLKSALGEKAYEWNDSEMYLFRSSVAYAMREYFSKFKK
U6DXQ3-1_N NSFVGWNTDWSPYADQSIKVRISLKSALGEKAYEWNDNEMYFFQSSIAYAMREYFSKVKK
BAE53380.1 NSFVGWNTDWSPYADQSIKVRISLKSALGEKAYEWNDNEMYFFQSSIAYAMREYFSKVKN
NP_0011587 NSFVGWNTDWSPYADQSIKVRISLKSALGEKAYEWNNNEMYLFRSSIAYAMRQYFSEVKN
AAX63775.1 NSFVGWDTDWRPYSDQSIKVRISLKSALGEKAYEWNDNEMYLFRSSIAYAMREYFSKVKN
XP_0070901 NSFVGWNTDWRPYADQSIKVRISLKSALGDKAYEWNDNEMYLFRSSVAYAMREYFSKVKN
NP_0010345 NSFVGWNTDWRPYADQSIKVRISLKSALGDEAYEWNDNEMYLFRSSVAYAMREYFSKVKN
:* ***.*:* **: ************** .**:*.:.*:::*.**:*****:** *:
AAW78017.1 QTVPFGEADVWVSDLKPRVSFNFFVTSPKNVSDIIPRSEVEEAIRMSRGRINDIFGLNDN
NP_0011239 QTVPFLEEDVRVSDLKPRVSFYFFVTSPQNVSDVIPRSEVEDAIRMSRGRINDVFGLNDN
AGZ48803.1 QTILFGAENVWVSNLKPRISFNFHVTSPGNLSDIIPRPEVEGAIRMSRSRINDAFRLDDN
NP_0011165 ETIPFGAVDVWVSDLKPRISFNFFVTSPANMSDIIPRSDVEKAISMSRSRINDAFRLDDN
AAY57872.1 QTILFGEEDVRVADLKPRISFNFYVTAPKNVSDIIPRTEVEEAIRFSRSRINDAFQLNDN
NP_0013583 QMILFGEEDVRVANLKPRISFNFFVTAPKNVSDIIPRTEVEKAIRMSRSRINDAFRLNDN
Q5RFN1.1_P QMILFGEEDVRVANLKPRISFNFFVTAPKNVSDIIPRTEVEKAIRMSRSRINDAFRLNDN
QLH93383.1 QTIPFEEESVRVSDLKPRVSFIFFVTLPKNVSAVIPRAEVEEAIRMSRSRINDVFRLDDN
U6DXQ3-1_N QTIPFVDKDVRVSDLKPRISFNFIVTSPENMSDIIPRADVEEAIRKSRGRINDAFRLDDN
BAE53380.1 QTIPFVGKDVRVSDLKPRISFNFIVTSPENMSDIIPRADVEEAIRKSRGRINDAFRLDDN
NP_0011587 QTIPFVEDNVWVSDLKPRISFNFSVTSPGNVSDIIPRTEVEEAIRMYRSRINDVFRLDDN
AAX63775.1 QTIPFVEDNVWVSDLKPRISFNFFVTFSNNVSDVIPRSEVEDAIRMSRSRINDAFRLDDN
XP_0070901 QTIPFVEDNVWVSNLKPRISFNFFVTASKNVSDVIPRREVEEAIRMSRSRINDAFRLDDN
NP_0010345 QTIPFVEDNVWVSNLKPRISFNFFVTASKNVSDVIPRSEVEEAIRMSRSRINDAFRLDDN
: : * .*.*::****:** * ** . *:* :*** :** ** *.**** * *:**
AAW78017.1 SLEFLGIYPTLKPPYEPPVTIWLIIFGVVMGTVVVGIVILIVTGIKGRKKKNETKREENP
NP_0011239 SLEFLGIHPTLEPPYQPPVTIWLIIFGVVMALVVVGIIILIVTGIKGRKKKNETKREENP
AGZ48803.1 SLEFLGIQPTLGPPYQPPVTIWLIVFGVVMAVVVVGIVVLIITGIRDRRKTDQARSEENP
NP_0011165 TLEFLGIQPTLGPPDEPPVTVWLIIFGVVMGLVVVGIVVLIFTGIRDRRKKKQASSEENP
AAY57872.1 SLEFLGIQSTLVPPYQSPITTWLIVFGVVMAVIVAGIVVLIFTGIRDRKKKNQARSEENP
NP_0013583 SLEFLGIQPTLGPPNQPPVSIWLIVFGVVMGVIVVGIVILIFTGIRDRKKKNKARSGENP
Q5RFN1.1_P SLEFLGIQPTLGPPNQPPVSIWLIVFGVVMGVIVVGIVVLIFTGIRDRKKKNKARNEENP
QLH93383.1 SLEFLGIQPTLEPPYQPPVTIWLIVFGVVMGVIVVGIVVLIFTGIRDRKKKNQARSEQNP
U6DXQ3-1_N SLEFLGIQPTLEPPYQPPVTIWLIVFGVVMGVVVVGIFLLIFSGIRNRRKNNQARSEENP
BAE53380.1 SLEFLGIQPTLEPPYQPPVTIWLIVFGVVMGVVVVGIFLLIFSGIRNRRKNNQARSEENP
NP_0011587 SLEFLGIQPTPGPPYEPPVTIWLIVFGVVMGVVVVGIVLLIFSGIRNRRKNDQARGEENP
AAX63775.1 SLEFLGIEPTLSPPYRPPVTIWLIVFGVVMGAIVVGIVLLIVSGIRNRRKNDQAGSEENP
XP_0070901 SLEFLGIQPTLSPPYQPPVTIWLIVFGVVMGVVVVGIVLLIVSGIRNRRKNNQARSEENP
NP_0010345 SLEFLGIQPTLSPPYQPPVTIWLIVFGVVMGVVVVGIVLLIVSGIRNRRKNNQARSEENP
:****** .* ** .*:: ***:*****. :*.**.:**.:**..*.*..:: :**
AAW78017.1 YDSMDIGKGESNAGFQNSDDAQTSF
NP_0011239 YDSMDIGKGESNAGFQNSDDAQTSF
AGZ48803.1 YSSVDLSKGENNPGFQNGDDVQTSF
NP_0011165 YGSMDLSKGESNSGFQNGDDIQTSF
AAY57872.1 YASIDISKGENNPGFQNTDDVQTSF
NP_0013583 YASIDISKGENNPGFQNTDDVQTSF
Q5RFN1.1_P YASIDISKGENNPGFQNTDDVQTSF
QLH93383.1 YASVDLSKGENNPGFQNVDDVQTSF
U6DXQ3-1_N YASVDLSKG----------------
BAE53380.1 YASVDLSKGENNPGFQNVDDVQTSF
NP_0011587 YASVDLSKGENNPGFQSGDDVQTSF
AAX63775.1 YASVDLNKGENNPGFQHADDVQTSF
XP_0070901 YASVDLSKGENNPGFQHADDVQTSF
NP_0010345 YASVDLSKGENNPGFQHADDVQTSF
* *:*:.**
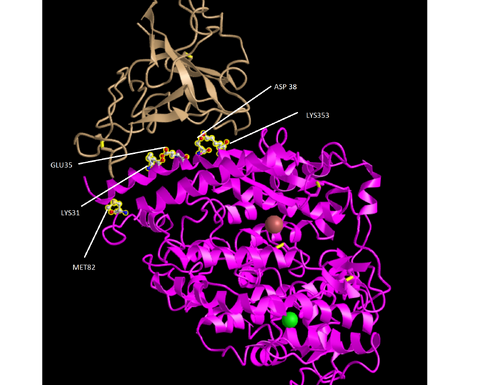
Model
To create the model we used the Figure 1C SARS-CoV RBD (optimized for human ACE2 recognition) and human ACE2: 3SCI. Follow the link to start the modelling process.
- Click on the Windows menu to “View Sequences & Annotations”
- In the new window that appears to the right, click on the “Details” tab to show the actual amino acid sequences
- There are 2 sets of ACE2-spike proteins because of the way the proteins crystallized.
- Focus on the pink and tan chains and orient them like is shown in Figure 4B
- We are going to make the amino acid side chains shown in the figure visible.
- In the sequence window go to sequence “Protein 3SCK_A” (in pink) and select the following amino acids. Use the overall
- K31
- E35
- D38
- M82
- K353
- The part of the ribbon that represents these amino acids should be highlighted in yellow in the structure
- Go to the Styles menu and select Proteins > Ball and Stick
- Go to the Color menu and select Atom
- You should see the side chains shown in the figure.
- The labels were then added using Microsoft Paint.
Phylogenetic tree
- My partner (Yaniv) created the phylogenetic tree using the FASTA sequencing and produced the phylogenetic tree on Phylogeny.fr

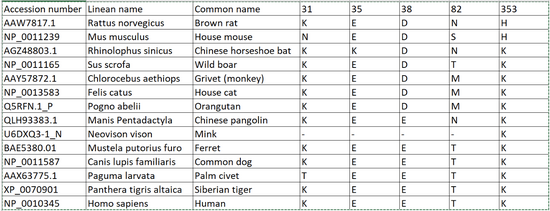
Species table
Using the Clustal format sequences we constructed the following table on Excel to better visualize the difference in the critical residues.
Presentation
File:Maddahi CorreyBioinformatics Presentation 1 slides.pdf
Conclusion
Overall this assignment taught me many things. I feel much more confident in my ability to do research and create models that assist with the research I am doing. We came to saveral conclusions about the binding of the ACE2 protein in different mammals with the SARS-COV-2 spike protein. The main conclusion we arrived at is that the binding depends on the polarity/charge of the amino acids in positions 31 and 353 in the ACE2 protein.
Acknowledgements
- Yaniv Maddahi
- Yaniv and I worked as homework partners for this week. We communicated and worked together both at the end of the week 6 lab and throughout the week to create our research project and assignment pages.
- Dr. Dahlquist
- Dr. Dahlquist served as a coach for how to begin our pages. She also instructed the class and provided us with the guiding homework document.
- I copied and modified the protocol shown on the Week 4 page of our class OpenWetWare.
- I copied and modified the protocol shown on the Week 5 page of our class OpenWetWare.
- I copied and modified sequence data and a phylogenetic tree from Phylogeny.fr
- I copied a model that was made using iCn3D web protein modelling.
Except for what is noted above, this individual journal entry was completed by me and not copied from another source. Jcorrey (talk) 23:02, 14 October 2020 (PDT)
References
- Yushun Wan, Jian Shang, Rachel Graham, Ralph S. Baric, Fang Li Journal of Virology Mar 2020, 94 (7) e00127-20; DOI: 10.1128/JVI.00127-20
- OpenWetWare. (2020). BIOL368/F20:Week 1. Retrieved September 30, 2020, from https://openwetware.org/wiki/BIOL368/F20:Week_1
- OpenWetWare. (2020). BIOL368/F20:Week 4. Retrieved September 30, 2020, from https://openwetware.org/wiki/BIOL368/F20:Week_4
- Phylogeny.fr: Home. (2020). Retrieved September 30, 2020, from https://www.phylogeny.fr/
- NCBI GenBank. (2020). Bat SARS coronavirus Rp3, complete genome - Nucleotide. Retrieved 1 October 2020, from https://www.ncbi.nlm.nih.gov/nuccore/DQ071615
- NCBI GenBank. (2020). spike protein [Bat SARS CoV Rp3/2004] - Protein. Retrieved 1 October 2020, https://www.ncbi.nlm.nih.gov/protein/72256271
- iCn3D: Web-based 3D Structure Viewer 2AJF. (2020). Retrieved 6 October 2020, from https://www.ncbi.nlm.nih.gov/Structure/icn3d/full.html?&mmdbid=35213&bu=1&showanno=1
- iCn3D: Web-based 3D Structure Viewer 3SCK. (2020). Retrieved 1 October 2020, from https://www.ncbi.nlm.nih.gov/Structure/icn3d/full.html?pdbid=%203SCK
- Uniprot. (2020). S - Spike glycoprotein precursor - Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) - S gene & protein. Retrieved 1 October 2020, from https://www.uniprot.org/uniprot/P0DTC2
- Andersen, K.G., Rambaut, A., Lipkin, W.I. et al. The proximal origin of SARS-CoV-2. Nat Med 26, 450–452 (2020). https://doi.org/10.1038/s41591-020-0820-9
JT Correy Template
Weekly Assignments
- Week 1 Assignment
- Week 2 Assignment
- Week 3 Assignment
- Week 4 Assignment
- Week 5 Assignment
- Week 6 Assignment
- Week 7 Assignment
- Week 8 Assignment
- Week 9 Assignment
- Week 10 Assignment
- Week 11 Assignment
- Week 12 Assignment
- Week 14 Assignment
Individual Journal Pages
- JT Correy
- JT Correy Journal Week 2
- JT Correy Journal Week 3
- JT Correy Journal Week 4
- JT Correy Journal Week 5
- JT Correy Journal Week 6
- JT Correy Journal Week 7
- CancerTracer Review
- JT Correy Journal Week 9
- JT Correy Journal Week 10
- JT Correy Journal Week 11
- JT Correy Journal Week 12
- JT Correy Journal Week 14
- The Mutants Week 14
Class Journal Pages
- BIOL368/F20:Class Journal Week 1
- BIOL368/F20:Class Journal Week 2
- BIOL368/F20:Class Journal Week 3
- BIOL368/F20:Class Journal Week 4
- BIOL368/F20:Class Journal Week 5
- BIOL368/F20:Class Journal Week 6
- BIOL368/F20:Class Journal Week 7
- BIOL368/F20:Class Journal Week 8
- BIOL368/F20:Class Journal Week 9
- BIOL368/F20:Class Journal Week 10
- BIOL368/F20:Class Journal Week 11
- BIOL368/F20:Class Journal Week 12
- BIOL368/F20:Class Journal Week 14