IGEM:Harvard/2006/DNA nanostructures/Container Design 3
Version 3.0
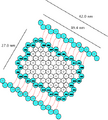
- This is a top view with top and bottom (lids) unattached.
- Each tube is 84 turns (or 27.5 nm) long.
- The pleated sheets connected to the box by red lines are cross-sectional views of the top and bottom (lids).
- The red lines represent short single-stranded linker sequences in the scaffold.
- Notice that the scaffold strand first enters a lid on a positive coil and leaves it on a negative coil (or vice versa).
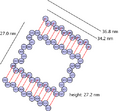
Dimensions
- 46 tubes (34.2 nm x 27.0 nm) x 84 bp strands (27.2 nm)
- each lid strand is connected to the adjacent barrel strand by a short single-strand linker
- each lid is 15 tubes wide (34.2 nm) x 84 bp (27.2 nm)
- total scaffold needed = 7116 bp
Version 3.1
- Structure is narrowed, ends are straightened
- Goal is easier and more regular lid attachment
- Schematic of DNA scaffold (version 3.1) shows linker oligo locations (blue) and possible aptamer binding sites (green)
- code and output
Dimensions
- 42 tubes (36.8 nm x 27.0 nm) x 84 bp strands (27.2 nm)
- 6132 bp needed
Version 3.2
Design changes
- lid linker sequences are 18 bp (4 bp previously)
- scaffold connector strands on lids are 24 bp (12 bp previously)
- one latch per lid (eight previously)
- latch connecting lid strands 014/015 to barrel strand 034
- latch connecting lid strands 033/034 to barrel strand 015
- 6612 bp needed
Top lid latch design
Linker (24 bp) between lid strand 014 and lid strand 015 (part of scaffold):
5'- GTAAATTCAGAGACTGCGCTTTCC -3'
3' end (21 bp) of scaffold of strand 034:
5'- GATGTTATTACTAATCAAAGA -3'
Scaffold-oligo-oligo-scaffold design:
5'- GGAAAGCGCAGT TTTT TTAGAATCAACCAG ATCGGTATGGCA -3' 5'- TCTTTGATTAG TTTT CTGGTTGATTCTAA TAGATGTGGCAA -3' Displacement strands (42 bp and 41 bp, respectively): 5'- TGCCATACCGATCTGGTTGATTCTAAAAAAACTGCGCTTTCC -3' 5'- TTGCCACATCTATTAGAATCAACCAGAAAACTAATCAAAGA -3'
Scaffold-oligo-staple-oligo-scaffold design:
5'- GGAAAGCGCAGT TTTT TTAGAATCAACCAG -3'
5'- TCGTGAAATTCTAA TGGCCCCAATAC -3'
5'- CTGGTTGGGATAGA GAGCTGTTCACC -3'
5'- TCTTTGATTAG TTTT TCTATCCTTCACGA -3'
Displacement strands (26 bp each):
5'- GTATTGGGGCCATTAGAATTTCACGA -3'
5'- GGTGAACAGCTCTCTATCCCAACCAG -3'
Old core oligo:
36 034 11 034 10 034 09 035 09 035 10 035 11
TCTTTGA TTAGTAA TAACATC TCCATCA CGCAAAT TAACCGT
New core oligo:
36* 034 10 034 09 035 09 035 10 035 11
TAA TAACATC TCCATCA CGCAAAT TAACCGT
Bottom lid latch design
Linker (24 bp) between lid strand 035 and lid strand 036 (part of scaffold):
5'- GCCCTGTAGCGGCGCATTAAGCGC -3'
3' end (21 bp) of scaffold of strand 013:
5'- ATCCGCCTGGTACTGAGCAAA -3'
Scaffold-oligo-oligo-scaffold design:
5'- GCGCTTAATGCG TTTT AGACGTAGTATCAC ATGCTCCACCCG -3' 5'- TGCGAATAGTACCA TTTT GTGATACTACGTCT GAGAAGATGTGC -3' Displacement strands (42 bp and 44 bp, respectively): 5'- CGGGTGGAGCATGTGATACTACGTCTAAAACGCATTAAGCGC -3' 5'- GCACATCTTCTCAGACGTAGTATCACAAAATGGTACTATTCGCA -3'
Scaffold-oligo-staple-oligo-scaffold design:
5'- GCGCTTAATGCG TTTT AGACGTAGTATCAC -3'
5'- TCCTGCATACGTCT ATGCTCCACCCG -3'
5'- GTGATACTATCTAT GAGAAGATGTGC -3'
5'- TGCGAATAGTACCA TTTT ATAGATATGCAGGA -3'
Displacement strands (26 bp each):
5'- CGGGTGGAGCATAGACGTATGCAGGA -3'
5'- GCACATCTTCTCATAGATAGTATCAC -3'
Old core oligo (42 bp):
1 010 01 011 01 011 02 012 02 012 01 013 01
ATTATAC CAAAAAA AAGGCTC AAGGAAT TGCGAAT AGTACCA
New core oligo (28 bp):
1* 010 01 011 01 011 02 012 02
ATTATAC CAAAAAA AAGGCTC AAGGAAT
Inward aptamers
Old oligo (42 bp):
1 013 08 014 08 014 07 015 07 015 08 016 08
GGAATAG GGAACCT ATTATTC ACCCTCA GAGCCAC TTTCATC
New oligo (60 bp):
1* 013 08 014 08 014 07 015 07 015 08 016 08
GGAATAG GGAACCT ATTATTC ACCCTCA GAGCCAC TTTCATC TTT GGTTGGTGTGGTTGG
Old oligo (42 bp):
2 036 07 035 07 035 08 034 08 034 07 033 07
AAGCACT AGTAAAA GAGTCTG ACTTGCC TGAGTAG ACAGAGG
New oligo (60 bp):
2* 036 07 035 07 035 08 034 08 034 07 033 07
AAGCACT AGTAAAA GAGTCTG ACTTGCC TGAGTAG ACAGAGG TTT GGTTGGTGTGGTTGG
Old oligo (42 bp):
1 004 03 004 02 004 01 005 01 005 02 005 03
AGTCAGA AGCAAAG CGGATTG GTAATAG TAAAATG TTTAGAC
New oligo (53 bp):
1* 004 03 004 02 004 01 005 01 005 02
AGTCAGA AGCAAAG CGGATTG GTAATAG TAAAATG TTT GGTTGGTGTGGTTGG
Outward apatmers
Old oligo (42 bp):
1 010 07 011 07 011 08 012 08 012 07 013 07
GGCAAAA ATCAGCT TGCTTTC TTTCAAC AGTTTCA ATAGCCC
New oligo (60 bp):
1* 010 07 011 07 011 08 012 08 012 07 013 07
GGCAAAA ATCAGCT TGCTTTC TTTCAAC AGTTTCA ATAGCCC TTT GGTTGGTGTGGTTGG
Old oligo (42 bp):
1 031 04 031 05 031 06 032 06 032 05 032 04
TCAGATG ATGGCAA TTCATCA CACCTTG CTGAACC TCAAATA
New oligo (53 bp):
1* 031 04 031 05 031 06 032 06 032 05
TCAGATG ATGGCAA TTCATCA CACCTTG CTGAACC TTT GGTTGGTGTGGTTGG
Old oligo (28 bp):
1 019 02 020 02 020 01 020 00
TTTATCC TCTTTCC AGAGCCT AATTTGC
New oligo (39 bp):
1* 019 02 020 02 020 01
TTTATCC TCTTTCC AGAGCCT TTT GGTTGGTGTGGTTGG
To do (old)
- reprogram oligos using my program — done
- use p8256 — decided not to use, 7308 is sufficient for now, has no insert
- check my program's output vs. OTP_ra (using Fifer's script) — spot-check is successful. thank you Fifer!
- spot-check oligos against "flowed" scaffold using InDesign — wrote script instead of using InDesign, but spot-checking successful, and caught bug in program
- make modifications on latch design — done
- finalize aptamer locations — done
- divide oligos into the following categories: — done
- see pre-working stocks below
- order oligos!
Matthewmeisel 12:51, 7 July 2006 (EDT)
Pre-working stocks
- c3.2.1: core barrel oligos
- c3.2.2: core top lid oligos
- c3.2.3: core bottom lid oligos
- c3.2.4: barrel oligos at inside apatamer locations -aptamers
- c3.2.5: barrel oligos at outside aptamer locations -aptamers
- c3.2.6: barrel oligos at inside apatamer locations +aptamers
- c3.2.7: barrel oligos at outside aptamer locations +aptamers
- c3.2.8: barrel oligos at latch locations -latches
- c3.2.9: lid oligos at latch locations -latches (empty)
- c3.2.10: barrel oligos at latch locations +latch1 +latch2
- c3.2.11: latch oligos from barrel +latch1 -latch2
- c3.2.12: latch oligos from barrel -latch1 +latch2
- c3.2.13: lid oligos at latch locations +latch1 +latch2 (empty)
- c3.2.14: latch oligos from lids +latch1 -latch2
- c3.2.15: latch oligos from lids -latch1 +latch2
- c3.2.16: latch staples +latch2
- c3.2.17: displacement strands: latch1
- c3.2.18: displacement strands: latch2
| Description | Contents | Total |
| c3.2.6L: barrel oligos @ inside aptamers + ligand | 3.2.6.1oa/ob, 3.2.6.2oa/ob, 3.2.6.3o | 5 |
| c3.2.7L: barrel oligos @ outside aptamers + ligand | 3.2.7.1oa/ob, 3.2.7.2o, 3.2.7.3o | 4 |
(mix 10 μL of each 50 μM stock ligand-oligos to make pre-working stocks)
Working stocks
list in progress
| core | aptamers | latches | latch displacement | |||||||||||||||||
| Stock ID | Experiment | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | |
| c3.2.A | -latches -aptamers | x | x | x | x | x | x | x | ||||||||||||
| c3.2.B | +latch1 -aptamers | x | x | x | x | x | x | x | x | x | ||||||||||
| c3.2.C | +latch2 -aptamers | x | x | x | x | x | x | x | x | x | x | |||||||||
| c3.2.D | -latches +aptamers_in | x | x | x | x | x | x | x | ||||||||||||
| c3.2.E | -latches +aptamers_out | x | x | x | x | x | x | x | ||||||||||||
| c3.2.F | +latch1 +aptamers_in | x | x | x | x | x | x | x | x | x | ||||||||||
| c3.2.G | +latch1 +aptamers_out | x | x | x | x | x | x | x | x | x | ||||||||||
| c3.2.H | +latch2 +aptamers_in | x | x | x | x | x | x | x | x | x | x | |||||||||
| c3.2.I | +latch2 +aptamers_out | x | x | x | x | x | x | x | x | x | x | |||||||||
| c3.2.J | displace latch1 | x | ||||||||||||||||||
| c3.2.K | displace latch2 | x | ||||||||||||||||||
| c3.2.L | -latches +aptamers_all | x | x | x | x | x | x | x | ||||||||||||
Final concentration of each oligo in working stocks is 250 nM.
| working stock | description | pre-working stocks (according to chart) | water | total |
| c3.2.E | -latches +aptamers_out |
1 (94 μL), 2 (28 μL), 3 (30 μL), 4 (3 μL), 7 (3 μL), 8 (2 μL), 9 (0 μL) | 40 μL | 200 μL |
| c3.2.Fo.core | +latch1 +aptamers_in |
1 (94 μL), 2 (28 μL), 3 (30 μL), 5 (3 μL), 6o (5 μL), 10 (2 μL), 11 (2 μL), 13 (0 μL) | 36 μL | 200 μL |
| c3.2.Fo.latches | +latch1 +aptamers_in |
14 (2 μL) | 10.5 μL | 12.5 μL of 4 μM |
| c3.2.Go.core | +latch1 +aptamers_out |
1 (94 μL), 2 (28 μL), 3 (30 μL), 4 (3 μL), 7o (4 μL), 10 (2 μL), 11 (2 μL), 13 (0 μL) | 37 μL | 200 μL |
| c3.2.Go.latches | +latch1 +aptamers_out |
14 (2 μL) | 10.5 μL | 12.5 μL of 4 μM |
| c3.2.Ho.core | +latch2 +aptamers_in |
1 (94 μL), 2 (28 μL), 3 (30 μL), 5 (3 μL), 6o (5 μL), 10 (2 μL), 12 (2 μL), 13 (0 μL), 15 (2 μL) | 34 μL | 200 μL |
| c3.2.Ho.latches | +latch2 +aptamers_in |
16 (4 μL) | 21 μL | 25 μL of 2 μM |
| c3.2.Io.core | +latch2 +aptamers_out |
1 (94 μL), 2 (28 μL), 3 (30 μL), 4 (3 μL), 7o (4 μL), 10 (2 μL), 12 (2 μL), 13 (0 μL), 15 (2 μL) | 35 μL | 200 μL |
| c3.2.Io.latches | +latch2 +aptamers_out |
16 (4 μL) | 21 μL | 25 μL of 2 μM |
| c3.2.Fo.all | +latch1 +aptamers_in |
1 (94 μL), 2 (28 μL), 3 (30 μL), 5 (3 μL), 6o (5 μL), 10 (2 μL), 11 (2 μL), 13 (0 μL), 14 (2 μL), oligo-ligand (1.5 μL 100 μM) | 32.5 μL | 200 μL |
| c3.2.Go.all | +latch1 +aptamers_out |
1 (94 μL), 2 (28 μL), 3 (30 μL), 4 (3 μL), 7o (4 μL), 10 (2 μL), 11 (2 μL), 13 (0 μL), 14 (2 μL), oligo-ligand (1.5 μL 100 μM) | 33.5 μL | 200 μL |
| c3.2.Ho.all | +latch2 +aptamers_in |
1 (94 μL), 2 (28 μL), 3 (30 μL), 5 (3 μL), 6o (5 μL), 10 (2 μL), 12 (2 μL), 13 (0 μL), 15 (2 μL), 16 (4 μL), oligo-ligand (1.5 μL 100 μM) | 28.5 μL | 200 μL |
| c3.2.Io.all | +latch2 +aptamers_out |
1 (94 μL), 2 (28 μL), 3 (30 μL), 4 (3 μL), 7o (4 μL), 10 (2 μL), 12 (2 μL), 13 (0 μL), 15 (2 μL), 16 (4 μL), oligo-ligand (1.5 μL 100 μM) | 29.5 μL | 200 μL |
Oligo list
EM images
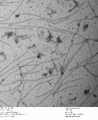
-
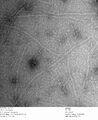
c3.2 -latches, 0.7% uranyl formate
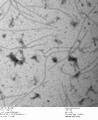
-
c3.2 -latches, 0.7% uranyl formate
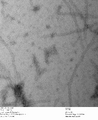
-
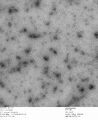
c3.2 +latch2, 0.7% uranyl formate
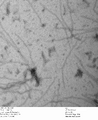
-
c3.2 +latch2, 0.7% uranyl formate
-
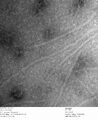
c3.2 +latch2, 0.23% uranyl formate
-
c3.2 +latch2, 0.23% uranyl formate
-
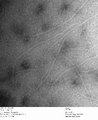
c3.2 +latch2, 0.23% uranyl formate
-
c3.2 +latch2, 0.23% uranyl formate
-
c3.2 +latch2, 0.23% uranyl formate
-
c3.2 +latch2, 0.23% uranyl formate
Oligo "ligand" aptamers
on the July 31 notebook page
Design figures
-
Version 3.0: top view, with cross sections of lids
-
Version 3.0: schematic of DNA scaffold
-
Version 3.1 top view, with cross sections of lids
-
Version 3.1: schematic of DNA scaffold (Illustrator file)
Notes about designs
- Calculating coil length: The distance between stacked base pairs is approximately (3.4 nm / turn) / (10.5 bp / turn) = 0.324 nm / bp.
- Calculating sheet length: The length of a "sheet" can be calculated using simple trigonometry, as demonstrated by the diagram below.
- The effective diameter of a coil is 3 nm according to Dr. Shih, but I'm not 100% sure that this figure is correct, so I will call it d until we can verify it.