IGEM:Harvard/2006/DNA nanostructures/Notebook/2006-7-31
Folding & Analysis of design 5
- Folding
- Mix the following, for each of .1 to .5 (v1 and v2):
- 4 uL 500 mM HEPES pH 7.5, 500 mM NaCl, 100 mM MgCl2
- 16 uL working stock 250 nM each oligo (100 nM each oligo final concentration)
- 20 uL p7392 20 nM (10 nM final concentration)
- Anneal from 80°C to 20°C, -1°C per min
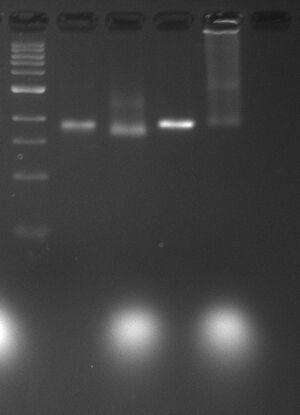
0.5x TBE, 11 mM MgCl2
| Lane | Contents | Loading Buffer |
| 0 | 1kb DNA ladder (4 μL) | |
| 1 | naked p7308 (12.5 μL) | AGLB (4 μL) |
| 2 | c5.0 barrel (12.5 μL) | AGLB (4 μL) |
| 3 | naked p7704 (12.5 μL) | AGLB (4 μL) |
| 4 | c5.0 lids (12.5 μL) | AGLB (4 μL) |
- Initial folding experiments of Design 5 barrel and lids
- Barrel
- Used p7308 scaffold for barrel, as designed
- Two bands are visible - one brighter band running at slightly faster mobility than naked scaffold, and a second more faint band running running slower, possibly indicating some dimization of the barrels.
- Lids
- Used p7704 scaffold because p7572 isn't yet available (should be able to make some by end of Tuesday
- significant amount of smearing is present, and high-molecular weight species are being retained at the top of the well
- two slight bands are visible within the smear (again may be monomeric and dimeric species), hopefully indicating that some percentage is properly folded
- will repeat with p7572 scaffold, possibly decrementing temperature -1°C per 2 min
Redesigning the DNA ligand
It turns out that NotI requires 10bp on each side of the restriction site for succesful digests. George also suggested that we include the restriction site on the double-stranded site of binding between the nanostructure and the oligo, because hairpins could constrain enzyme binding.
We need to redesign the old DNA ligand.
Restriction site choice
Other possible restriction enzymes are listed below. Verified that restriction site does not exist in p7308 using "find" function in Notepad. Minimum number of bp on each side is from NEB chart. Molecular weights and chain lengths are from NEB product technical support (1-800-632-7799).
- AflIII (ACATGT)
- sequence not present in p7308
- >90% yield after 2 hr digest with 2 bp on each side
- 26,954 Da (233 aa)
- AscIII (GGCGCGCC)
- sequence not present in p7308
- >90% yield after 2 hr digest with 0 bp on each side
- 50,466 Da (455 aa)
- StuI (AGGCCT)
- sequence not present in p7308
- >90% yield after 2 hr digest with 1 bp on each side
- 28,518 Da (255 aa)
None of these enzymes has confirmed star activity.
It's advantageous to choose the largest enzyme (least likely to fit into the nanostructure), so we choose AscIII.
Schematic
Random generation script
A sample output:
TAAAGAAACTCGGATGCGCCACGCAGGATTGGGGCGCGCCCGCGG
The script flanks the restriction site with random G-C pairs for stronger Watson-Crick base pairing. Because of the short length (15 bp) of the overlap, it is impossible to introduce another AscIII restriction site, so the script does not check for the additional site. The script was adopted from an earlier version:
#!/usr/bin/python
import random
import sys
import string
BamHI = 'ggatcc'
EcoRI = 'gaattc'
NotI = 'gcggccgc'
AscI = 'ggcgcgcc'
aptamer = 'GGTTGGTGTGGTTGG'
def rev(s):
return s[::-1]
complement = string.maketrans('ACGTacgt','TGCAtgca')
def comp(s):
return rev(s.translate(complement))
def hasresite(s):
result = False
if s.count(BamHI) > 0:
print 'BamHI found'
result = True
elif s.count(EcoRI) > 0:
print 'EcoRI found'
result = True
elif s.count(NotI) > 0:
print 'NotI found'
result = True
elif s.count(AscI) > 0:
print 'NotI found'
result = True
return result
# prints random sequence, length specified as argument
def randseq(l):
flag = True
s = []
while (flag == True):
s = []
for i in range(l):
s.append(random.choice(['a', 'c', 'g', 't']))
flag = hasresite(''.join(s))
return ''.join(s)
def randseqCG(l):
flag = True
s = []
while (flag == True):
s = []
for i in range(l):
s.append(random.choice(['c', 'g']))
flag = hasresite(''.join(s))
return ''.join(s)
#if len(sys.argv) > 1:
# a = int(sys.argv[1])
#else:
# sys.exit("usage: ./random-sequence.py [length]")
final_seq = []
##oligo design 1
#final_seq.append(randseq(39))
#final_seq.append(NotI)
#final_seq.append('tttt')
#final_seq.append(NotI)
#final_seq.append('tttt')
#final_seq.append(comp(aptamer))
## oligo design 2
final_seq.append(randseq(30))
final_seq.append(randseqCG(2))
final_seq.append(AscI)
final_seq.append(randseqCG(5))
final_seq = ''.join(final_seq).upper()
print final_seq
Redesigning the "aptamers"
Previously, we had designed ligand oligos that were complimentary to the thrombin aptamer, which meant we could use the existing aptamer sequences. For this new design, however, we need to order new "aptamer" sequences.
Five more thymine nucleotides were added, too, so the enzyme won't have to act as close to the nanostructure.
Old 3.2 aptamer sequences:
c3.2.6.1 GGAATAGGGAACCTATTATTCACCCTCAGAGCCACTTTCATCTTTGGTTGGTGTGGTTGG c3.2.6.2 AAGCACTAGTAAAAGAGTCTGACTTGCCTGAGTAGACAGAGGTTTGGTTGGTGTGGTTGG c3.2.6.3 AGTCAGAAGCAAAGCGGATTGGTAATAGTAAAATGTTTGGTTGGTGTGGTTGG c3.2.7.1 GGCAAAAATCAGCTTGCTTTCTTTCAACAGTTTCAATAGCCCTTTGGTTGGTGTGGTTGG c3.2.7.2 TCAGATGATGGCAATTCATCACACCTTGCTGAACCTTTGGTTGGTGTGGTTGG c3.2.7.3 TTTATCCTCTTTCCAGAGCCTTTTGGTTGGTGTGGTTGG
New 3.2 aptamer sequences:
c3.2.6.1oa GGAATAGGGAACCTATTATTCACCCTCA c3.2.6.1ob GAGCCACTTTCATCTTTTTTTTCCGCGGGCGCGCCCC c3.2.6.2oa AAGCACTAGTAAAAGAGTCTGACTTGCC c3.2.6.2ob TGAGTAGACAGAGGTTTTTTTTCCGCGGGCGCGCCCC c3.2.6.3o AGTCAGAAGCAAAGCGGATTGGTAATAGTAAAATGTTTTTTTTCCGCGGGCGCGCCCC c3.2.7.1oa GGCAAAAATCAGCTTGCTTTCTTTCAAC c3.2.7.1ob AGTTTCAATAGCCCTTTTTTTTCCGCGGGCGCGCCCC c3.2.7.2o TCAGATGATGGCAATTCATCACACCTTGCTGAACCTTTTTTTTCCGCGGGCGCGCCCC c3.2.7.3o TTTATCCTCTTTCCAGAGCCTTTTTTTTTCCGCGGGCGCGCCCC
New 5.0 Oligo Ligand "Aptamers": (xxx indicates the aptamer exit point)
NB: These oligos are listed by their original oligo number (ie. "oligo 7"). The first oligo (with the xxx in it) in the list is the original oligo, with the spaces showing where the oligo should be split; the second and third oligos are the ones that will be ordered - these are split in order to guarantee accuracy of the sequence (impeded if the oligo is greater than 60nt long).
- Outside @ Aptamers
oligo 7 c5.0.4.1 ATAAATCGTGTTGTTCCAGTT xxx TGGAACAAGAGTCCGTAAAGC ATAAATCGTGTTGTTCCAG TTTTTTTTTT CCGCGGGCGCGCCCC TGGAACAAGAGTCCGTAAAGC oligo 43 c5.0.4.2 GCCGGAAGCAGGTCGACTCTAAGGGGGA xxx TGTGCTGCGGAAAC GCCGGAAGCAGGTCGACTCTAAGGGGGA TTTTTTTT CCGCGGGCGCGCCCC TGTGCTGCGGAAAC oligo 65 c5.0.4.3 TCATTTTTTTACAAACAATTCAAATGAA AAATCTAGATAAAA xxx TCATTTTTTTACAAACAATTCAAATGAA AAATCTAGATAAAA TTTTTTTT CCGCGGGCGCGCCCC oligo 75 c5.0.4.4 ATCAATAGATAAAAATTTTTA GAACCCTCATATATATTAGCA xxx ATCAATAGATAAAAATTTTTA GAACCCTCATATATATTAGCA TTTTTTTT CCGCGGGCGCGCCCC oligo 78 c5.0.4.5 GGTATTAAATATCCCATCCTAATTT xxx ACGAGCATGTCGAGCCA GGTATTAAATATCCCATCCTAATTT TTTTTTTT CCGCGGGCGCGCCCC ACGAGCATGTCGAGCCA
- Inside @ Aptamers
oligo 33 c5.0.5.1 GCCGCCATTGATATTCACAAA xxx CAAATAAATCCTCATCTGAAT GCCGCCATTGATATTCACAAA TTTTTTTT CCGCGGGCGCGCCCC CAAATAAATCCTCATCTGAAT oligo 67 c5.0.5.2 ATTTACATTTTGCGGGATCGTGAAGTTT CCATTAA xxx ACGGGTA ATTTACATTTTGCGGGATCGTGAAGTTT CCATTAA TTTTTTTT CCGCGGGCGCGCCCC oligo 89 c5.0.5.3 TGGATAGCGATAAAAACCAAA xxx ATAGCGAGAGGCTTACAACAT TGGATAGCGATAAAAACCAAA TTTTTTTT CCGCGGGCGCGCCCC ATAGCGAGAGGCTTACAACAT oligo 95 c5.0.5.4 AAAGAAAAATGAATTTTCTGTTCACCAG TACAAAC xxx TACAACG AAAGAAAAATGAATTTTCTGTTCACCAG TACAAAC TTTTTTTT CCGCGGGCGCGCCCC oligo 104 c5.0.5.5 TCATTTGTTCTGCG xxx AACGAGTGGTCATTTTTGCGGACCAGAC TCATTTGTTCTGCG TTTTTTTT CCGCGGGCGCGCCCC AACGAGTGGTCATTTTTGCGGACCAGAC
PEG precipitations
Used PEG precipitation protocol for reagents shown for lanes 5-20. Pellet lanes: reconstituted pellet in 10 μL folding buffer and loaded it all. Supernatant lanes: loaded 10 μL supernatant.
Some lanes were not loaded. 6hb not loaded b/c we had no folded 6hb. 10%, 12%, 14% supernatant not loaded so that everything could fit on one gel.

| lane | folding reaction (μL) | 20% peg / 2.5 M NaCl (μL) | water (μL) | final PEG concentration | pellet/supernatant |
| 1 | 1 kb+ ladder | ||||
| 2 | 10 | 0 | 0 | 0% | |
| x | 10 (6hb) | 20 | 10 | 10% | pellet |
| x | 10 (6hb) | 20 | 10 | 10% | supernatant |
| 3 | 10 | 12 | 18 | 6% | pellet |
| 4 | 10 | 12 | 18 | 6% | supernatant |
| 5 | 10 | 16 | 14 | 8% | pellet |
| 6 | 10 | 16 | 14 | 8% | supernatant |
| 7 | 10 | 20 | 10 | 10% | pellet |
| x | 10 | 20 | 10 | 10% | supernatant |
| 8 | 10 | 22 | 8 | 11% | pellet |
| 9 | 10 | 22 | 8 | 11% | supernatant |
| 10 | 10 | 24 | 6 | 12% | pellet |
| x | 10 | 24 | 6 | 12% | supernatant |
| 11 | 10 | 26 | 4 | 13% | pellet |
| 12 | 10 | 26 | 4 | 13% | supernatant |
| 13 | 10 | 28 | 2 | 14% | pellet |
| x | 10 | 28 | 2 | 14% | supernatant |
| 14 | 10 | 30 | 0 | 15% | pellet |
| 15 | 10 | 30 | 0 | 15% | supernatant |
Results/discussion:
- appears to be good yields at 10% and above
- only supernatant lane with oligos visible is lane 6
- no pellet lanes contain oligo smears (but oligo smears difficult to see in supernatant lanes)
- difficult to see oligos (maybe run on PA gel next time?)
- where are the expected oligo smears in higher percent supernatants?
- unclear why unprecipitated nanostructures (lane 2) ran faster
- we're using structures that were folded at least a week ago, and some have been stored on the bench -- will try again tomorrow with nanostructures that are being folded today
- dye in supernatant lanes ran a little slow -- PEG or NaCl interference?
Gel purification of nanostructures
- using new buffer. added to the buffer 1 mL of 1M MgCl2 per 100 mL buffer
- using 20ul of folded structure.
- trying both design 3 and design 4
- using scaffold as a control (to hopefully varify we're cutting out nanostructures), but we know roughly where the nanostructures run to in any case.
Ordered Latches for c5.0
NB: Sent order in and was denied - short by four oligos. Will try to add four tomorrow and resend. Added c3.2 and c5.0 attachment oligos for oligo-ligand design; order accepted. c4.0 attachment oligos were ordered separately.
Order form for v5.0 latches, v3.2 attachment oligos, and v5.0 attachment oligos
c5.0.10: Barrel Latches
c5.0.11: Barrel Splits (ie. portion of oligo that had to be split off from the latch to allow addition of latch sequence )
c5.0.12: Barrel Zipper Oligos
c5.0.13: Top Lid Latches
c5.0.14: Top Lid Splits
c5.0.15: Top Lid Zipper Oligos
c5.0.16: Bottom Lid Latches
c5.0.17: Bottom Lid Splits
c5.0.18: Bottom Lid Zipper Oligos
c5.0.19: Barrel Displacement Latches
c5.0.20: Top Lid Displacement Latches
c5.0.21: Bottom Lid Displacement Latches
High Concentrations of Nanostructure Experiment, Day 1
- Mixed un-divided (ie. core and latches in same tube) working stocks of Gb (biotinylated oligos outside, with latches1) and Ib (biotinylated oligos outside, with latches2).
- Folded reactions of Gb and Eb, 8 40uL reactions each, overnight. Wanted to fold Ib and Ab, but ran out of p7308.