Dcartmel Week 13
User Page Link
Template Page Link
Assignment Pages
Individual Journal Pages
- Dcartmel Week 2
- Dcartmel Week 3
- Dcartmel Week 4
- Dcartmel Week 5
- Dcartmel Week 6
- Dcartmel Week 8
- Dcartmel Week 9
- Dcartmel Week 10
- Dcartmel Week 11
- Dcartmel Week 13
- Dcartmel Week 14
Shared Class Journal Pages
- Class Journal Week 1
- Class Journal Week 2
- Class Journal Week 3
- Class Journal Week 4
- Class Journal Week 5
- Class Journal Week 6
- Class Journal Week 7
- Class Journal Week 8
- Class Journal Week 9
- Class Journal Week 10
- Class Journal Week 13
- Class Journal Week 14
Purpose
The purpose of this assignment is to become familiar with different resources that can allow us to visualize the molecular structure of spike proteins and other related proteins.
Combined Methods/Results
- Converted the spike protein DNA sequence into a protein sequence using ExPASY Translate tool
- How do you know which of the six frames is the correct reading frame (without looking up the answer)?
- To determine which of the six frames is the correct reading frame, you need to look for the one that contains the most highlighted sequence. In this case, the first reading frame was the correct reading frame because it has the most amount of highlighting.
- Once you answered the question above, you can check your answer with the NCBI protein record.
- Checked for the correct answer. The correct reading frame is Frame 1.
- How do you know which of the six frames is the correct reading frame (without looking up the answer)?
- Find out what is already known about the spike protein in the UniProt Knowledgebase (UniProt KB).
- If you search on the keywords "SARS-CoV" (which refers to the first SARS virus), in the main UniProt search field, how many results do you get?
- By searching the keywords "SARS-CoV", I got 833 results.
- Used entry with accession number "P59594" which corresponds to the reference entry for the SARS-CoV spike protein.
- What types of information are provided about this protein in this database entry?
- The types of information that are provided about this protein in this database entry are its function, names and taxonomy, subcellualr locations, information on the diseases and phenotypes associated with the protein, post translational modifications, how it interacts with other proteins, information about teritary and secondary structure, sequence similarities it shares with other proteins and the domains that it has, the sequence of the protein, and links to proteins that are similar.
- If you search on the keywords "SARS-CoV" (which refers to the first SARS virus), in the main UniProt search field, how many results do you get?
- Use the PredictProtein server to analyze the SARS-CoV-2 spike protein.
- Pasted the SARS-CoV spike protein amino acid sequence discussed in journal club paper Wan et al. (2020) into the input field.
- How does this information relate to what is stored in the UniProt database for the SARS-CoV spike protein?
- This information relates to what is stored in the UniProt database for the SARS-CoV protein because both databases provide descriptions/predictions of the structure of the proteins as well as information about the sequences and sequence alignments.
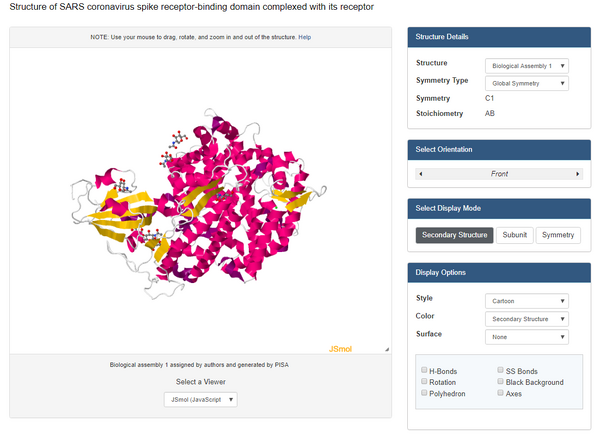
- View the structure of the SARS-CoV or SARS-CoV-2 spike protein from your assigned journal club article at the Protein Data Bank.
- Used this link for Wan data:Wan et al. (2020): 2AJF
- Clicked on the "Structure" underneath the image of the structure on the upper left side of the page.
- At the bottom right of the screen, you will see a drop-down menu that says "Select a different viewer". Select "JSMol" or "NGL" to access a palette of options for viewing the structure
- Alternately, NCBI has a web-based structure viewer called iCn3D
- Go to the NCBI Structure home page and paste the PDB ID for your structure into the search field and click "search".
- On the results page, you will see an image of the protein structure. Click on the button for the "full-featured 3D viewer" found on the bottom right corner of the structure image.
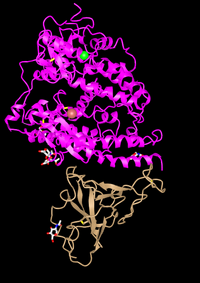
- Create a view of the protein from your paper recreates one of the figures from your paper. You may not be able to get this to be exactly the same in terms of colors or backbone style, but you should try to rotate the to be the same view as the article.
- Structure from original (Wan et al, 2020) paper:
- Alternately, NCBI has a web-based structure viewer called iCn3D
- Recreated structure:
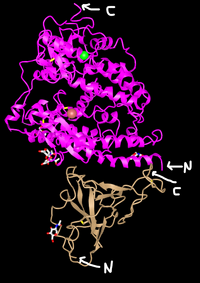
- N-Terminus and C-Terminus labeled in each tertiary structure:
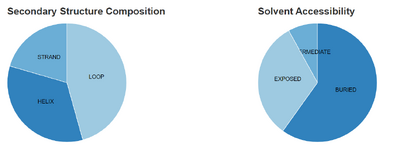
- The secondary structures in the recreated figure do match with the predictions that were made in the Predict Protein database.
- Secondary structure proportions from Predict Protein database
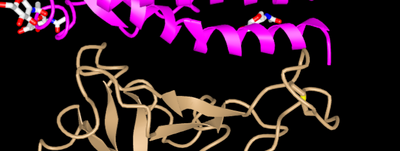
- The Wan et al (2020) paper discussed amino acids 442, 472, 479, 486, and 487 on the SARS-human virus and amino acids 455, 486 493. 494, and 501 on the 2019nCoV virus.
- In this picture, the amino acids 442, 472, 479,486 and 487 of the SARS-human virus are located on the golden colored structure in the bottom of the picture.
Beginning your research project
With your new final project partners, answer the following:
- What question will you answer about sequence-->structure-->function relationships in a SARS-CoV-2 protein?
- In the Yan et al. (2020) paper, RBD-PD complexes of SARS-CoV and SARS-CoV-2 were compared and it was found that differences in residues lead to weaker interactions. Walls et al. (2020) discusses that when the SARS outbreak of 2002 reemerged in 2003-04, the virus had a weaker interaction with ACE2 and patients showed milder symptoms. This research project will further explore th role that certain amino acids play in the structure-function relationship of SARS-CoV-2 and ACE2.
- What sequences will you use? I want you to take advantage of sequence data available to perform a multiple sequence alignment as part of your project.
- ACE2 sequences from humans, mice, and bats will be used.
- We will be using humans because humans are currently dealing this this virus.
- We will use mice because they have been unaffected by the virus possibly because of the interaction between ACE2 and the S protein.
- We will use bats because the virus is thought to have started in bats and then had been transmitted to humans.
- ACE2 sequences from humans, mice, and bats will be used.
- What protein tools will you use for analysis and answering your question?
- We will use sequences from UniPort to conduct a multiple sequence alignment. The 3-D structures will be compared using iCn3D and other programs with hopes to see if there are any structural differences that will weaken or inhibit the binding to the S protein.
Data and Files
- Media:Week 13 DC Prob 1.png
- Media:Week 13 DC Prob 3.png
- Media:Week 13 DC Prob 4 .png
- Media:Week 13 actual structure from paper DC.png
- Media:Week 13 recreated structure .png
- Media:Week 13 recreated structure labeled .png
- Media:Week 13 structure comparison.png
- Media:Week 13 amino acids.png
Scientific Conclusion
The overall goal of this assignment was to become familiar with different available tools that can allow us to analyze the 3-D structure of viral proteins. The results of this assignment showed that there is agreement between sources on the overall structure and characteristics of the corona virus.
Acknowledgements
I worked with my homework partners Jack P. Menzagopian and Nicholas D. Yeo. We texted each other on 4/22/2020 to work on the research project questions.
I copied and modified the Week 13 assignment page.
Except for what is noted above, this individual journal entry was completed my me and not copied from another source.
Dcartmel (talk) 22:50, 22 April 2020 (PDT)
References
OpenWetWare. (2020). BIOL368/S20:Week 13. Retireved April 22, 2020 from https://openwetware.org/wiki/BIOL368/S20:Week_13.
ExPASY. (2020) ExPASy Translate Tool. Retrieve April 22, 2020 from http://web.expasy.org/translate/ ExPASY Translate tool.
UniProt. (2020). Retrieved April 22, 2020 from http://www.uniprot.org UniProt.
NCBI. (2020). Structure Database. Retrieved April 22, 2020 from https://www.ncbi.nlm.nih.gov/Structure/index.shtml NCBI Structure Database.
NCBI. (2020). NCBI Open Reading Frame Finder, Retrieved April 22, 2020 from https://www.ncbi.nlm.nih.gov/orffinder/ NCBI Open Reading Frame Finder.
Wan, Y., Shang, J., Graham, R., Baric, R. S., & Li, F. (2020). Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. Journal of virology, 94(7). Retrieved April 22, 2020 from https://jvi.asm.org/content/94/7/e00127-20.