Carolyne week 14
Purpose
The purpose of this investigation is to determine how the changes in structure of non-human primate ACE2 compared to human ACE2 may affect interactions between SARS-CoV-2 and its target receptor ACE2.
Methods/Results
Finding Relevant Residues
- I retrieved the FASTA protein sequences for the following primate species from UniProtKB from searching "ace2": Rhinopithecus bieti, Macaca nemestrina, Homo sapiens, Aotus nancymaae , Saimiri boliviensis boliviensis, Macaca mulatta, Propithecus coquereli, Tarsius syrichta, Rhinopithecus roxellana, Papio anubis, Macaca fascicularis, Chlorocebus sabaeus, Nomascus leucogenys, Pan paniscus, Pongo abelii, Pan troglodytes
- First, I aligned the human, sumatran orangutan (Sumat), chimpanzee (Chimp), and bonobo (Bonob) ACE2 sequences together to see if they have residue differences in the areas highlighted by Yan et. al. I chose these species because the latter 3 are very closely related to humans. The results of the alignment are displayed below:
CLUSTAL FORMAT: MUSCLE (3.8) multiple sequence alignment
ACE2_Sumat MSGSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLASWNYNTNITEENVQ
ACE2_Human MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLASWNYNTNITEENVQ
ACE2_Chimp MSGSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLASWNYNTNITEENVQ
ACE2_Bonob MSGSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLASWNYNTNITEENVQ
**.*********************************************************
ACE2_Sumat NMNNAGDKWSAFLKEQSTLAQMYPLQEIQNLTVKLQLQALQQNGSSVLSEDKSKRLNTIL
ACE2_Human NMNNAGDKWSAFLKEQSTLAQMYPLQEIQNLTVKLQLQALQQNGSSVLSEDKSKRLNTIL
ACE2_Chimp NMNNAGDKWSAFLKEQSTLAQMYPLQEIQNLTVKLQLQALQQNGSSVLSEDKSKRLNTIL
ACE2_Bonob NMNNAGDKWSAFLKEQSTLAQMYPLQEIQNLTVKLQLQALQQNGSSVLSEDKSKRLNTIL
************************************************************
ACE2_Sumat NTMSTIYSTGKVCNPNNPQECLLLEPGLNEIMANSLDYNERLWAWESWRSEVGKQLRPLY
ACE2_Human NTMSTIYSTGKVCNPDNPQECLLLEPGLNEIMANSLDYNERLWAWESWRSEVGKQLRPLY
ACE2_Chimp NTMSAIYSTGKVCNPNNPQECLLLEPGLNEIMANSLDYNERLWAWESWRSEVGKQLRPLY
ACE2_Bonob NTMSAIYSTGKVCNPNNPQECLLLEPGLNEIMANSLDYNERLWAWESWRSEVGKQLRPLY
****:**********:********************************************
ACE2_Sumat EEYVVLKNEMARANHYEDYGDYWRGDYEVNGVDSYDYSRGQLIEDVEHTFEEIKPLYEHL
ACE2_Human EEYVVLKNEMARANHYEDYGDYWRGDYEVNGVDGYDYSRGQLIEDVEHTFEEIKPLYEHL
ACE2_Chimp EEYVVLKNEMARANHYEDYGDYWRGDYEVNGVDGYDYSRGQLIEDVEHTFEEIKPLYEHL
ACE2_Bonob EEYVVLKNEMARANHYEDYGDYWRGNYEVNGVDGYDYSRGQLIEDVEHTFEEIKPLYEHL
*************************:*******.**************************
ACE2_Sumat HAYVRAKLINAYPSYISPIGCLPAHLLGDMWGRFWTNLYSLTVPFGQKPNIDVTDAMVDQ
ACE2_Human HAYVRAKLMNAYPSYISPIGCLPAHLLGDMWGRFWTNLYSLTVPFGQKPNIDVTDAMVDQ
ACE2_Chimp HAYVRAKLMNAYPSYISPIGCLPAHLLGDMWGRFWTNLYSLTVPFGQKPNIDVTDAMVDQ
ACE2_Bonob HAYVRAKLMNAYPSYISPIGCLPAHLLGDMWGRFWTNLYSLTVPFGQKPNIDVTDAMVDQ
********:***************************************************
ACE2_Sumat AWDAQRIFKEAEKFFVSVGLPNMTQRFWENSMLTDPGNVQKVVCHPTAWDLGKGDFRILM
ACE2_Human AWDAQRIFKEAEKFFVSVGLPNMTQGFWENSMLTDPGNVQKAVCHPTAWDLGKGDFRILM
ACE2_Chimp AWDAQRIFKEAEKFFVSVGLPNMTQGFWENSMLTDPGNVQKAVCHPTAWDLGKGDFRILM
ACE2_Bonob AWDAQRIFKEAEKFFVSVGLPNMTQGFWENSMLTDPGNVQKAVCHPTAWDLGKGDFRILM
************************* ***************.******************
ACE2_Sumat CTKVTMDDFLTAHHEMGHIQYDMAYAAQPFLLRNGANEGFHEAVGEIMSLSAATPKHLKS
ACE2_Human CTKVTMDDFLTAHHEMGHIQYDMAYAAQPFLLRNGANEGFHEAVGEIMSLSAATPKHLKS
ACE2_Chimp CTKVTMDDFLTAHHEMGHIQYDMAYAAQPFLLRNGANEGFHEAVGEIMSLSAATPKHLKS
ACE2_Bonob CTKVTMDDFLTAHHEMGHIQYDMAYAAQPFLLRNGANEGFHEAVGEIMSLSAATPKHLKS
************************************************************
ACE2_Sumat IGLLSPDFQEDNETEINFLLKQALTIVGTLPFTYMLEKWRWMVFKGEIPKDQWMKKWWEM
ACE2_Human IGLLSPDFQEDNETEINFLLKQALTIVGTLPFTYMLEKWRWMVFKGEIPKDQWMKKWWEM
ACE2_Chimp IGLLSPDFQEDNETEINFLLKQALTIVGTLPFTYMLEKWRWMVFKGEIPEDQWMKKWWEM
ACE2_Bonob IGLLSPDFQEDNETEINFLLKQALTIVGTLPFTYMLEKWRWMVFKGEIPKDQWMKKWWEM
*************************************************:**********
ACE2_Sumat KREIVGVVEPVPHDETYCDPASLFHVSNDYSFIRYYTRTLYQFQFQEALCQAAKHEGPLH
ACE2_Human KREIVGVVEPVPHDETYCDPASLFHVSNDYSFIRYYTRTLYQFQFQEALCQAAKHEGPLH
ACE2_Chimp KREIVGVVEPVPHDETYCDPASLFHVSNDYSFIRYYTRTLYQFQFQEALCQAAKHEGPLH
ACE2_Bonob KREIVGVVEPVPHDETYCDPASLFHVSNDYSFIRYYTRTLYQFQFQEALCQAAKHEGPLH
************************************************************
ACE2_Sumat KCDISNSTEAGQKLLNMLRLGKSEPWTLALENVVGAKNMNVRPLLDYFEPLFTWLKDQNK
ACE2_Human KCDISNSTEAGQKLFNMLRLGKSEPWTLALENVVGAKNMNVRPLLNYFEPLFTWLKDQNK
ACE2_Chimp KCDISNSTEAGQKLFNMLRLGKSEPWTLALENVVGAKNMNVRPLLNYFEPLFTWLKDQNK
ACE2_Bonob KCDISNSTEAGQKLFNMLRLGKSEPWTLALENVVGAKNMNVRPLLNYFEPLFTWLKDQNK
**************:******************************:**************
ACE2_Sumat NSFVGWSTDWSPYADQSIKVRISLKSALGNKAYEWNDNEIYLFRSSVAYAMRKYFLEVKN
ACE2_Human NSFVGWSTDWSPYADQSIKVRISLKSALGDKAYEWNDNEMYLFRSSVAYAMRQYFLKVKN
ACE2_Chimp NSFVGWSTDWSPYADQSIKVRISLKSALGDKAYEWNDNEMYLFRSSVAYAMRQYFLKVKN
ACE2_Bonob NSFVGWSTDWSPYADQSIKVRISLKSALGDKAYEWNDNEMYLFRSSVAYAMRQYFLKVKN
*****************************:*********:************:***:***
ACE2_Sumat QMILFGEEDVRVANLKPRISFNFFVTAPKNVSDIIPRTEVEKAIRMSRSRINDAFRLNDN
ACE2_Human QMILFGEEDVRVANLKPRISFNFFVTAPKNVSDIIPRTEVEKAIRMSRSRINDAFRLNDN
ACE2_Chimp QMILFGEEDVRVANLKPRISFNFFVTAPKNVSDIIPRTEVEKAIRKSRSRINDAFRLNDN
ACE2_Bonob QMILFGEEDVRVANLKPRISFNFFVTAPKNVSDIIPRTEVEKAIRKSRSRINDAFRLNDN
********************************************* **************
ACE2_Sumat SLEFLGIQPTLGPPNQPPVSIWLIVFGVVMGVIVVGIVVLIFTGIRDRKKKNKARNEENP
ACE2_Human SLEFLGIQPTLGPPNQPPVSIWLIVFGVVMGVIVVGIVILIFTGIRDRKKKNKARSGENP
ACE2_Chimp SLEFLGIQPTLGPPNQPPVSIWLIVFGVVMGVIVVGIVILIFTGIRDRKKKNKARSEENP
ACE2_Bonob SLEFLGIQPTLGPPNQPPVSIWLIVFGVVMGVIVVGIVILIFTGIRDRKKKNKARSEENP
**************************************:****************. ***
ACE2_Sumat YASIDISKGENNPGFQNTDDVQTSF
ACE2_Human YASIDISKGENNPGFQNTDDVQTSF
ACE2_Chimp YASVDTSKGENNPGFQNTDDVQTSF
ACE2_Bonob YASVDTSKGENNPGFQNTDDVQTSF
***:* *******************
- Then, I copied the aligned sequences into Word, and used the sequences highlighted in Figure 4 of the Yan et. al. (2020) paper to determine if all of the sequences were the same by highlighting the relevant residues in yellow.
- From this analysis, I found that all of these species had the same residues in the same locations highlighted by Yan et. al. (2020)
- I then went ahead to align the other primate species ACE2 sequences to human ACE2 to see if any of the primates had unique changes at the relevant positions.
- RESULTS SUMMARY: Many of the primates had sequences aligned and identical to the sequences seen at the critical spots in human ACE2. However, some species had alterations at the following positions identified in the Yan et. al. (2020) publication: Y41, Q42, K353, and R357.
Visualizng ACE2 and SARS-COV-2
- I found a protein structure visualization and manipulation program called UCSF Chimera. I downloaded the program on to my computer and did the following to visualize the hACE2 structure and SARS-CoV-2:
- Go to File -> Fetch by ID… -> entered 6M0J (the ID to attain the SARS-CoV-2 with hACE2 pictured in Yan et. al. (2020)
- The protein structure appeared in the program.
- First, I wanted to change the color to better distinguish SARS-CoV-2 from hACE2
- To select the SARS-CoV-2 go to Select-> Chain -> A. hACE2 should be selected in green.
- To change the color, go to Actions -> Color -> . hACE2 became light sea green
- To stop selecting hACE2, go to Select -> Clear selection
- Next, I wanted to view the actual residues that the Yan et. al. paper highlighted.
- To select the individual residues, I went to Select -> Atom specifier... -> ::(Single-Letter residue abbreviation):(Position) (ex. ::Y:41)
- The residue would become outlined in green to show it was selected.
- To select multiple residues, I held down the shift and control buttons, then clicked the desired residues with my mouse
- I selected the residues highlighted in the Yan et. al. paper, then changed the colors of the important residues to yellow in ACE2 and green in SARS-Cov-2.
- For all residues I looked at, I showed the heteroatomic coloration by selecting the residues and going to Actions -> Color -> by heteroatom
- Then I wanted to determine how the interactions would change if the ACE2 residues in hACE2 were altered to reflect the sequences in primates that had sequence differences. First I wanted to create the Y41H alteration that is seen in S. bolivensis bolivensis
- I held control key, then click to select residue Y41 in ACE2
- With Y41 selected, I went to Tools -> Stucture Editing -> Rotamers
- A window should pop up. Then, I went to Rotamer type -> HIS (H is the single-letter abbreviation for histidine, HIS is the 3 letter abbreviation for histidine). Then I selected Rotamer library -> Dunbrack 2010. Once those parameters were selected, I clicked apply.
- A new window should pop up with the probabilities listed for the possible rotamers. I selected the rotamer with the highest probability (this will be the one listed first usually). I went to existing side chain -> retain to keep the original residue visible. Then I clicked "apply"
- This should show the original side chain and the new residue alteration. Then I clicked "Ok" and the window closed.
- I held down the control key and clicked on the altered side chain. I changed the color by going to Action -> Color -> orange then Action -> Color-> by heteroatom
- To change the residues for Q42E, K353L, and R347D, I repeated this process. The Y41H and Q42E alterations were done together to reflect the structure of S. bolivensis bolivensis. the K353L and R357D alterations were done together to reflect the structure of R. bieti.
- Finally, I wanted to superimpose the normal hACE2 structure on top of the altered structure I created so that the location changes of the altered amino acids could be easily seen.
- I went to File -> Open...
- A new window popped up. I selected the file "" which had the normal hACE2 structure
- The new window closed, but a popup appeared asking "". Click "no"
- The result should be that the 2 hACE2 SARS-CoV-2 models are superimposed on each other since most of the resiudes are in the same position. The only residues that should not be the same are the Rotamer alterations, which will be out of place and in orange.
- To focus on the amino acids mentioned by Yan et. al. (2020), I held down both the control and shift keys so I could select multiple amino acids. I clicked all of the orange, yellow, and green residues.
- With the desired residues selected, I went to Actions -> Focus. The window should now focus on the selected residues, with most of the extra ribbon from SARS-CoV-2 and hACE2 not in sight
- If there was extra ribbon blocking my view, I held down shift and control, and I clicked on the ribbon I wanted to remove to select those parts. Then I went to Actions -> Atoms/Bonds -> hide. With the same parts still selected, I went to Actions -> Ribbons -> hide. These two operations should remove ribbons that block the view of the amino acids and was repeated to remove additional ribbons until I had a good view of the residue alterations.
- I rotated and zoomed in and out of the view until I had a good view of the original hACE2 residues (yellow), altered hACE2 residues (orange), and SARS-CoV-2 residues (green)
- To save the window image as a .PNG image, I went to File -> Save Image... and then saved the image with the desired name in a folder (but it can be saved anywhere)
- To save the sesion, go to File -> Save Session or File -> Save Session As...
- After doing these steps to show the alterations in R. bieti and S. bolivensis bolivensis, I went to File -> Close session and then exited out of Chimera by closing the program
Primate Alteration Molecular Models
- Saimiri boliviensis boliviensis and Aotus nancymaae: Y41H and Q42E
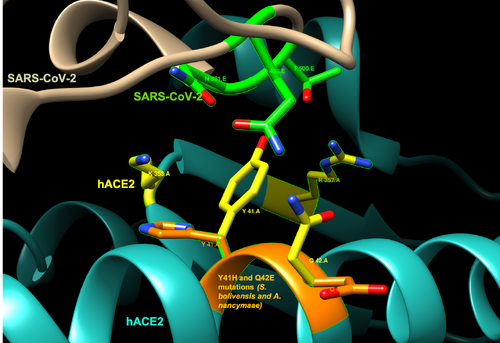
Both the Y41H and Q42E alterations seen in S. boliviensis and A. nancymaae position the amino acids in positions that are not optimal for interaction with the SARS-CoV-2. In addition, the alteration Y41H changes a nonpolar aromatic side chain (tyrosine) chain to a positive side chain (histidine), while the alteration Q42E changes an uncharged polar side chain (glutamine) to a negatively charged side chain (glutamate).
- Rhinopithecus bieti: R357D and K353L alterations
Both the R357D and K353L alterations seen in R. bieti position the amino acids in positions that are not optimal for interaction with the SARS-CoV-2. In addition, the alteration R357D changes a positive side chain (arginine) chain to a negative side chain (aspartate), while the alteration K353L changes a positive side chain (lysine) to a non-polar side chain (leucine).
Scientific Conclusion
The purpose of this investigation was to determine how structural differences in the ACE2 receptor of human and non-human primates may affect how SARS-CoV-2 interacts with the receptor in the species. The residues examined were the nine residues of ACE2 featured in Figure 4 of the Yan et. al. (2020) paper. Overall, as expected, at the positions highlighted by Yan et. al. (2020) many non-human primate ACE2 structures have the same 9 residues observed in human ACE2. However, in primates that do have pairs of alterations at these positions, the altered residues may drastically lead to changes in interactions with residues 498, 500, and 501 in SARS-CoV-2. These pairs of altered residues (Y41H and Q42E; R357D and K353L) are not only positioned away from the relevant SARS-CoV-2 residues but also have different chemical properties. Based on this work, more research and modelling should be done to see if these alterations lead to greater structural changes that make non-human primates less or more likely to be vulnerable to SARS-CoV-2.
Data and Files
- Box Link (including alignments, presentation, and UCSF Chimera Files): https://lmu.box.com/s/dm7n9ihxy8i01k77xg2h0etkhjl6o6os
- Link to image files on OpenWetWare: Media:Ver 2 Y41H and Q42E non-human primate mutants.png, Media:Ver 2 R357D and K353L non-human primate mutants.png
- Presentation Link: https://docs.google.com/presentation/d/1dgE04--rlMtsh5fluO4TE9X7tNcga9NXxMLrm_gxz3o/edit?usp=sharing
Acknowledgements
I would like to acknowledge my homework partners Jenny and Nathan. We met over Zoom last week and talked several times over text during this past week to work on the project, create the presentation, and figure out how to divide the workload. I copied the Yan et. al. (2020) citation from the Week 13 Assignment page and used the instructions from the Week 14 Assignment page to organize my assignment page. I created the images of the protein structures using UCSF Chimera and use the help guide they provided to create the images shown here. I aligned the amino acid sequences with Phylogeny.fr. I retrieved the amino acid sequences for ACE2 from UniProtKB. I used the syntax from the Wikipedia help page for pictures to help me format the images on my assignment page. Except for what is noted above, this individual journal entry was completed by me and not copied from another source.Carolyne (talk) 10:07, 30 April 2020 (PDT)
References
- UniProt (2020). UniProtKB results "ace2". Retrieved April 23, 2020 from https://www.uniprot.org/uniprot/?query=ace2&sort=score.
- Phylogeny.fr (2020). Phylogeny.fr Robust Phylogenetic Analysis For The Non-Specialist. Retrieved April 29, 2020 from http://www.phylogeny.fr/.
- Yan, R., Zhang, Y., Li, Y., Xia, L., Guo, Y., & Zhou, Q. (2020). Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science, 367(6485), 1444-1448. doi: 10.1126/science.abb2762
- Shapovalov, M. V., & Dunbrack, R. L., Jr. (2011). A smoothed backbone-dependent rotamer library for proteins derived from adaptive kernel density estimates and regressions. Structure (London, England : 1993), 19(6), 844-858. doi:10.1016/j.str.2011.03.019
- Pettersen, E. F., Goddard, T.D., Huang, C.C., Couch, G.S., Greenblatt, D.M., Meng, E.C., & Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004 Oct;25(13):1605-12.
- Resource for Biocomputing, Visualization, and Informatics (RBVI). (2018). UCSF Chimera: An extensible molecular modeling system. Retrieved April 29, 2020 from https://www.cgl.ucsf.edu/chimera/.
- OpenWetWare. (2020). BIOL368/S20:Week 13. Retrieved April 30, 2020, from https://openwetware.org/wiki/BIOL368/S20:Week_13.
- OpenWetWare. (2020). BIOL368/S20:Week 14. Retrieved April 23, 2020, from https://openwetware.org/wiki/BIOL368/S20:Week_13.
- Wikipedia. (2020). Help:Pictures. Retrieved April 30, 2020 from https://en.wikipedia.org/wiki/Help:Pictures
User Page and Template Links
- User page: Carolyn C. Egekeze
- Template: Template:Carolyne
Individual Journal Pages
- Carolyne week 2
- Carolyne week 3
- Carolyne week 4
- Carolyne week 5
- Carolyne week 6
- Carolyne week 8
- Carolyne week 9
- Carolyne week 10
- Carolyne week 11
- Carolyne week 13
- Carolyne week 14
Weekly Assignments
- BIOL368/S20:Week 1
- BIOL368/S20:Week 2
- BIOL368/S20:Week 3
- BIOL368/S20:Week 4
- BIOL368/S20:Week 5
- BIOL368/S20:Week 6
- BIOL368/S20:Week 8
- BIOL368/S20:Week 9
- BIOL368/S20:Week 10
- BIOL368/S20:Week 11
- BIOL368/S20:Week 13
- BIOL368/S20:Week 14
Class Journal Pages
- BIOL368/S20:Class Journal Week 1
- BIOL368/S20:Class Journal Week 2
- BIOL368/S20:Class Journal Week 3
- BIOL368/S20:Class Journal Week 4
- BIOL368/S20:Class Journal Week 5
- BIOL368/S20:Class Journal Week 6
- BIOL368/S20:Class Journal Week 8
- BIOL368/S20:Class Journal Week 9
- BIOL368/S20:Class Journal Week 10
- BIOL368/S20:Class Journal Week 11
- BIOL368/S20:Class Journal Week 13
- BIOL368/S20:Class Journal Week 14