Carolyne week 13
Purpose
The purpose of this activity is to investigate the protein structure of SARS-CoV-2's spike protein to learn more about how the protein sequence can give rise to the structure.
Methods/Results
SARS-CoV-2 Spike protein ORF
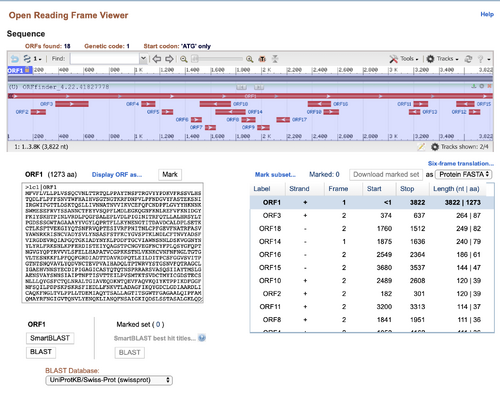
- I pasted the DNA sequence of the SARS-CoV-2 spike protein into the NCBI ORF Finder
- The correct reading frame would probably be ORF 1 because that produces the longest protein. Since the spike-protein is fairly large, it would require many amino acids to compose the structure.
- After checking the NCBI protein record, I found my answer was correct.
SARS-CoV UniProt Records
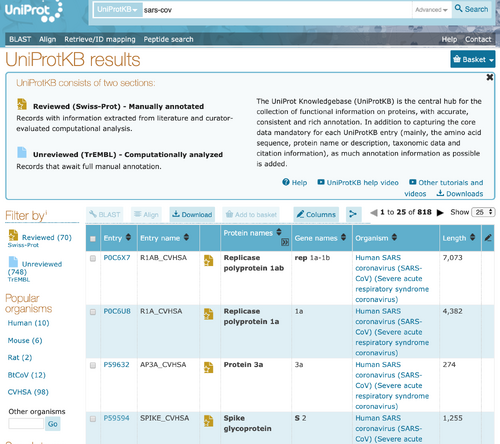
- I went to UniProt KB and searched for "SARS-CoV" in the search bar near the top of the page.
- The search returned 818 results, containing results from both Swis-Prot and TrEMBL. I clicked on the result that had the accession number "P59594".
- In this database entry, there are many types of information provided. The entry displays the protein, gene, and organism that the spike glycoprotein comes from. It contains information regarding the function of spike proteins 1 and 2. The entry also contains information on the subcellular localization, pathology, structure, interactions, and sequence the protein has. The structure information, in particular, includes a 3D image of the protein and information about the location of the secondary structures. Finally, it also tells the user what proteins are similar to the SARS-CoV glycoprotein and other databases that may have information about the protein as well.
Analyzing SARS-CoV-2 Spike Protein
PredictProtein Results
- I went to the Supplementary Materials file for the Yan et. al. (2020) paper. The authors included the Uniprot ID for the spike protein of SARS-CoV-2 they isolated. I went to UniProt, entered the ID in the search bar, and clicked on the search result with the ID. I scrolled down to the "sequence" section and copied the protein amino acid sequence for the SARS-CoV-2 spike protein.
- I went to PredictProtein and pasted the amino acid sequence into the input box and clicked PredictProtein.
- I received the following result below:
- The types of information provided by PredictProtein primarily focus on the structure and function of the protein. Regarding structure, PredictProtein includes information about the predicted protein disorder, secondary structures are predicted, residues accessibility to solvents. It also contains information to regarding predictions of disulfide bridges and transmembrane helices. Regarding function, PredictProtein includes information about subcellular localization, binding sites, and impacts of point mutations. They also have predictions regarding the biological processes and molecular functions the protein is a part of (gene ontology).
- Compared to the UniProt entry on SARS-CoV, the information gained from PredictProtein seems cover many of the same topics when it comes to structure and function. Both sites have information on the possible functions of the protein, the biological processes the protein is involved in, where it is found in the cell, the binding sites on the protein. They also both have information about the sequence and secondary structures of the protein. However, PredictProtein only had information regarding structure and function predictions. It did not have any information or predictions relating to where the protein sequence came from and how the protein might be processed. Finally, another big difference is that the UniProt entry for SARS-CoV spike protein included a 3D model of the structure. Since PredictProtein is making predictions, there is no 3D structure model provided.
Yan et. al. (2020) SARS-CoV-2 structure
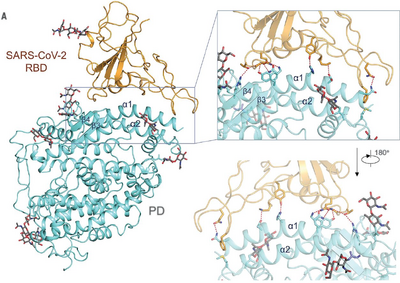
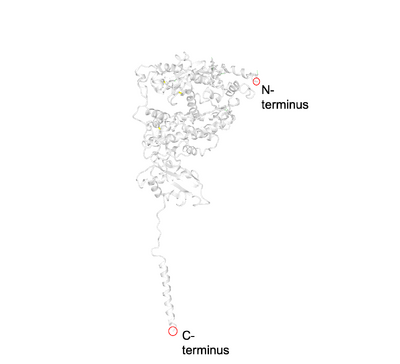
- I clicked the link for the Yan et. al. (2020) structure on the Week 13 assignment page to view the structure at the Protein Data Bank. For reference, the structure's PDB code is 6M17. Then I used the NCBI's iCn3D viewer to view the structure. I went to the NCBI structure page, entered the PDB code into the search bar, and then used the full-featured 3D viewer to view the protein.
- I did my best to try and recreate Figure 4A from the Yan et. al. (2020) paper. I've included both the image from the paper and the image that I made to recreate the figure.
- The following images show the locations of the C-terminus and N-terminus in SARS-CoV-2 and ACE2.
- The PredictProtein program predicted that the SARS-CoV-2 would have primarily loops, and about an even amount of helices and sheets. From the structure attained by Yan et. al. (2020), it appears that SARS-CoV-2 has about 8 sheets that are connected by loops, but no helices. Thus the protein prediction was somewhat accurate, but not completely accurate.
- This image shows the amino acids of SARS-CoV 2 that were important for binding to ACE2.
Scientific Conclusion
The aim of this activity was to study SARS-CoV-2's spike protein to understand how protein sequence can give rise to the structure and function. While it is unclear exactly how the protein folds based on its sequence alone, the sequence can help with predicting the likely secondary structures. Moreover, the PredictProtein site showed that there is a lot that can be learned or predicted about the function of the protein based on its structure alone. By using databases and examining the structure of the protein in 3D, the relationship between structure and function becomes more clear.
Data and Files
Coronavirus Project
- For the coronavirus project, my group would like to study the qualities (structure, genome,etc.) that make ACE2 a good receptor for SARS-CoV-2. We plan to use the spike protein sequence from SARS-CoV-2.
Acknowledgements
I copied and modified my methods/results section based on the outline that was in the Week 13 assignment page. I also copied the Yan et. al. (2020) citation from the Week 13 assignment page. I copied the syntax to embed my images from Wikipedia. I talked with Nathan and Jenny to over Zoom and over text to discuss ideas for the final project. Except for what is noted above, this individual journal entry was completed by me and not copied from another source.Carolyne (talk) 21:49, 22 April 2020 (PDT)
References
- Yan, R., Zhang, Y., Li, Y., Xia, L., Guo, Y., & Zhou, Q. (2020). Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science, 367(6485), 1444-1448. doi: 10.1126/science.abb2762
- OpenWetWare. (2020). BIOL368/S20:Week 13. Retrieved April 16, 2020, from https://openwetware.org/wiki/BIOL368/S20:Week_13.
- Wikipedia. (2020). Help:Pictures. Retrieved April 22, 2020 from https://en.wikipedia.org/wiki/Help:Pictures
User Page and Template Links
- User page: Carolyn C. Egekeze
- Template: Template:Carolyne
Individual Journal Pages
- Carolyne week 2
- Carolyne week 3
- Carolyne week 4
- Carolyne week 5
- Carolyne week 6
- Carolyne week 8
- Carolyne week 9
- Carolyne week 10
- Carolyne week 11
- Carolyne week 13
- Carolyne week 14
Weekly Assignments
- BIOL368/S20:Week 1
- BIOL368/S20:Week 2
- BIOL368/S20:Week 3
- BIOL368/S20:Week 4
- BIOL368/S20:Week 5
- BIOL368/S20:Week 6
- BIOL368/S20:Week 8
- BIOL368/S20:Week 9
- BIOL368/S20:Week 10
- BIOL368/S20:Week 11
- BIOL368/S20:Week 13
- BIOL368/S20:Week 14
Class Journal Pages
- BIOL368/S20:Class Journal Week 1
- BIOL368/S20:Class Journal Week 2
- BIOL368/S20:Class Journal Week 3
- BIOL368/S20:Class Journal Week 4
- BIOL368/S20:Class Journal Week 5
- BIOL368/S20:Class Journal Week 6
- BIOL368/S20:Class Journal Week 8
- BIOL368/S20:Class Journal Week 9
- BIOL368/S20:Class Journal Week 10
- BIOL368/S20:Class Journal Week 11
- BIOL368/S20:Class Journal Week 13
- BIOL368/S20:Class Journal Week 14