BioMicroCenter:PREP TEST
| HOME -- | SEQUENCING -- | LIBRARY PREP -- | HIGH-THROUGHPUT -- | COMPUTING -- | DATA MANAGEMENT -- | OTHER TECHNOLOGY |
Library Preparation Services
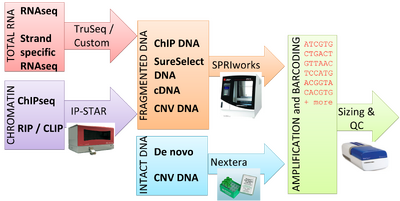
Generating high-quality data on the Illumina sequencing platform requires high-quality libraries. The BMC currently offers library preparation services for a variety of starting materials. Prior to sequencing, all samples must pass the BioMicro Center’s Sequencing Quality Control process, which verifies selection of inserts of a desired size and correct ligation of Illumina adapters.
Illumina Library Prep Services
Please follow the links in the table below for more information about our library preparation offerings.
| METHOD TYPE | DNA | RNA |
|---|---|---|
| STANDARD | Ligation-mediated PCR Methods
Tagmentation Methods |
Standard Input
Low Input
Small RNA upon request |
| HIGH THROUGHPUT | Ligation-mediated PCR Methods
Tagmentation Methods
Amplicon Methods
|
Standard Input
Low Input
|
OTHER SERVICES
SIZE SELECTION
The BMC offers PippinPrep Size Selection post library construction if custom insert sizes are needed.
QUALITY CONTROL
All NGS libraries are quality controlled prior to use. The BioMicro Center offers two 'flavors' of quality control for Illumina libraries.
Standard quality control includes analysis on the Fragment Analyzer and quantification by four point qPCR quantification for each sample. This confirms both the sizing as well as the presence of Illumina adapters on the libraries.
Rapid Quality Control is less expensive and quicker. It includes analysis on the Fragment Analyzer and quantification by PicoGreen on the Varioskan for each sample. Final pools are quantified by qPCR before loading. While this method is significantly faster and less expensive, we cannot guarantee pool balance with it.
