Anna Horvath Week 6
From OpenWetWare
Jump to navigationJump to search
Purpose
To determine, through a comparison of eighteen different ACE-2 sequences, which of them are likely intermediary host for SARS-CoV-2, which has the most similar ACE2 receptor to humans. This could then be used to inform future comparisons of coronaviruses by understanding the ACE-2 similarities
Methods/Results
Part 1: GenBank
- I went to GenBank in order to find the nineteen sequences.
- Sequences used included:
- humans [2]
- civet [3]
- Chinese bats [4]
- mice [5]
- rats [6]
- pigs [7]
- ferrets [8]
- cats [9]
- orangutans [10]
- grivet monkeys [11]
- fox [12]
- chickens [13]
- king cobras [14]
- pangolins [15]
- dromedary camels [16]
- squirrels [17]
- mink [18]
- Chinese softshell turtles [19]
>NP_001358344.1 angiotensin-converting enzyme 2 isoform 1 precursor [Homo sapiens] MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLASWNYNTNITEENVQNMNNAGDKWS AFLKEQSTLAQMYPLQEIQNLTVKLQLQALQQNGSSVLSEDKSKRLNTILNTMSTIYSTGKVCNPDNPQE CLLLEPGLNEIMANSLDYNERLWAWESWRSEVGKQLRPLYEEYVVLKNEMARANHYEDYGDYWRGDYEVN GVDGYDYSRGQLIEDVEHTFEEIKPLYEHLHAYVRAKLMNAYPSYISPIGCLPAHLLGDMWGRFWTNLYS LTVPFGQKPNIDVTDAMVDQAWDAQRIFKEAEKFFVSVGLPNMTQGFWENSMLTDPGNVQKAVCHPTAWD LGKGDFRILMCTKVTMDDFLTAHHEMGHIQYDMAYAAQPFLLRNGANEGFHEAVGEIMSLSAATPKHLKS IGLLSPDFQEDNETEINFLLKQALTIVGTLPFTYMLEKWRWMVFKGEIPKDQWMKKWWEMKREIVGVVEP VPHDETYCDPASLFHVSNDYSFIRYYTRTLYQFQFQEALCQAAKHEGPLHKCDISNSTEAGQKLFNMLRL GKSEPWTLALENVVGAKNMNVRPLLNYFEPLFTWLKDQNKNSFVGWSTDWSPYADQSIKVRISLKSALGD KAYEWNDNEMYLFRSSVAYAMRQYFLKVKNQMILFGEEDVRVANLKPRISFNFFVTAPKNVSDIIPRTEV EKAIRMSRSRINDAFRLNDNSLEFLGIQPTLGPPNQPPVSIWLIVFGVVMGVIVVGIVILIFTGIRDRKK KNKARSGENPYASIDISKGENNPGFQNTDDVQTSF
>AAX63775.1 angiotensin-converting enzyme 2 [Paguma larvata] MSGSFWLLLSFAALTAAQSTTEELAKTFLETFNYEAQELSYQSSVASWNYNTNITDENAKNMNEAGAKWS AYYEEQSKLAQTYPLAEIQDAKIKRQLQALQQSGSSVLSADKSQRLNTILNAMSTIYSTGKACNPNNPQE CLLLEPGLDNIMENSKDYNERLWAWEGWRAEVGKQLRPLYEEYVALKNEMARANNYEDYGDYWRGDYEEE WTGGYNYSRNQLIQDVEDTFEQIKPLYQHLHAYVRAKLMDTYPSRISRTGCLPAHLLGDMWGRFWTNLYP LTVPFGQKPNIDVTDAMVNQNWDARRIFKEAEKFFVSVGLPNMTQGFWENSMLTEPGDGRKVVCHPTAWD LGKGDFRIKMCTKVTMDDFLTAHHEMGHIQYDMAYAAQPFLLRNGANEGFHEAVGEIMSLSAATPNHLKT IGLLSPAFSEDNETEINFLLKQALTIVGTLPFTYMLEKWRWMVFKGAIPKEQWMQKWWEMKRNIVGVVEP VPHDETYCDPASLFHVANDYSFIRYYTRTIYQFQFQEALCQIAKHEGPLHKCDISNSTEAGKKLLEMLSL GRSEPWTLALERVVGAKNMNVTPLLNYFEPLFTWLKEQNRNSFVGWDTDWRPYSDQSIKVRISLKSALGE KAYEWNDNEMYLFRSSIAYAMREYFSKVKNQTIPFVEDNVWVSDLKPRISFNFFVTFSNNVSDVIPRSEV EDAIRMSRSRINDAFRLDDNSLEFLGIEPTLSPPYRPPVTIWLIVFGVVMGAIVVGIVLLIVSGIRNRRK NDQAGSEENPYASVDLNKGENNPGFQHADDVQTSF
>AGZ48803.1 angiotensin-converting enzyme 2 [Rhinolophus sinicus] MSGSSWLLLSLVAVTTAQSTTEDEAKMFLDKFNTKAEDLSHQSSLASWDYNTNINDENVQKMDEAGAKWS AFYEEQSKLAKNYSLEQIQNVTVKLQLQILQQSGSPVLSEDKSKRLNSILNAMSTIYSTGKVCKPNKPQE CLLLEPGLDNIMGTSKDYNERLWAWEGWRAEVGKQLRPLYEEYVVLKNEMARGYHYEDYGDYWRRDYETE ESPGPGYSRDQLMKDVERIFTEIKPLYEHLHAYVRAKLMDTYPFHISPTGCLPAHLLGDMWGRFWTNLYP LTVPFGQKPNIDVTDEMLKQGWDADRIFKEAEKFFVSVGLPNMTEGFWNNSMLTEPGDGRKVVCHPTAWD LGKGDFRIKMCTKVTMEDFLTAHHEMGHIQYDMAYASQPYLLRNGANEGFHEAVGEVMSLSVATPKHLKT MGLLSPDFREDNETEINFLLKQALNIVGTLPFTYMLEKWRWMVFKGEIPKEEWMKKWWEMKRKIVGVVEP VPHDETYCDPASLFHVANDYSFIRYYTRTIFEFQFHEALCRIAQHDGPLHKCDISNSTDAGKKLHQMLSV GKSQAWTKTLEDIVDSRNMDVGPLLKYFEPLYTWLQEQNRKSYVGWNTDWSPYSDQSIKVRISLKSALGE NAYEWNDNEMYLFRSSVAYAMREYFLKEKHQTILFGAENVWVSNLKPRISFNFHVTSPGNLSDIIPRPEV EGAIRMSRSRINDAFRLDDNSLEFLGIQPTLGPPYQPPVTIWLIVFGVVMAVVVVGIVVLIITGIRDRRK TDQARSEENPYSSVDLSKGENNPGFQNGDDVQTSF
>NP_001123985.1 angiotensin-converting enzyme 2 precursor [Mus musculus] MSSSSWLLLSLVAVTTAQSLTEENAKTFLNNFNQEAEDLSYQSSLASWNYNTNITEENAQKMSEAAAKWS AFYEEQSKTAQSFSLQEIQTPIIKRQLQALQQSGSSALSADKNKQLNTILNTMSTIYSTGKVCNPKNPQE CLLLEPGLDEIMATSTDYNSRLWAWEGWRAEVGKQLRPLYEEYVVLKNEMARANNYNDYGDYWRGDYEAE GADGYNYNRNQLIEDVERTFAEIKPLYEHLHAYVRRKLMDTYPSYISPTGCLPAHLLGDMWGRFWTNLYP LTVPFAQKPNIDVTDAMMNQGWDAERIFQEAEKFFVSVGLPHMTQGFWANSMLTEPADGRKVVCHPTAWD LGHGDFRIKMCTKVTMDNFLTAHHEMGHIQYDMAYARQPFLLRNGANEGFHEAVGEIMSLSAATPKHLKS IGLLPSDFQEDSETEINFLLKQALTIVGTLPFTYMLEKWRWMVFRGEIPKEQWMKKWWEMKREIVGVVEP LPHDETYCDPASLFHVSNDYSFIRYYTRTIYQFQFQEALCQAAKYNGSLHKCDISNSTEAGQKLLKMLSL GNSEPWTKALENVVGARNMDVKPLLNYFQPLFDWLKEQNRNSFVGWNTEWSPYADQSIKVRISLKSALGA NAYEWTNNEMFLFRSSVAYAMRKYFSIIKNQTVPFLEEDVRVSDLKPRVSFYFFVTSPQNVSDVIPRSEV EDAIRMSRGRINDVFGLNDNSLEFLGIHPTLEPPYQPPVTIWLIIFGVVMALVVVGIIILIVTGIKGRKK KNETKREENPYDSMDIGKGESNAGFQNSDDAQTSF
>AAW78017.1 angiotensin converting enzyme 2 [Rattus norvegicus] MSSSCWLLLSLVAVATAQSLIEEKAESFLNKFNQEAEDLSYQSSLASWNYNTNITEENAQKMNEAAAKWS AFYEEQSKIAQNFSLQEIQNATIKRQLKALQQSGSSALSPDKNKQLNTILNTMSTIYSTGKVCNSMNPQE CFLLEPGLDEIMATSTDYNRRLWAWEGWRAEVGKQLRPLYEEYVVLKNEMARANNYEDYGDYWRGDYEAE GVEGYNYNRNQLIEDVENTFKEIKPLYEQLHAYVRTKLMEVYPSYISPTGCLPAHLLGDMWGRFWTNLYP LTTPFLQKPNIDVTDAMVNQSWDAERIFKEAEKFFVSVGLPQMTPGFWTNSMLTEPGDDRKVVCHPTAWD LGHGDFRIKMCTKVTMDNFLTAHHEMGHIQYDMAYAKQPFLLRNGANEGFHEAVGEIMSLSAATPKHLKS IGLLPSNFQEDNETEINFLLKQALTIVGTLPFTYMLEKWRWMVFQDKIPREQWTKKWWEMKREIVGVVEP LPHDETYCDPASLFHVSNDYSFIRYYTRTIYQFQFQEALCQAAKHDGPLHKCDISNSTEAGQKLLNMLSL GNSGPWTLALENVVGSRNMDVKPLLNYFQPLFVWLKEQNRNSTVGWSTDWSPYADQSIKVRISLKSALGK NAYEWTDNEMYLFRSSVAYAMREYFSREKNQTVPFGEADVWVSDLKPRVSFNFFVTSPKNVSDIIPRSEV EEAIRMSRGRINDIFGLNDNSLEFLGIYPTLKPPYEPPVTIWLIIFGVVMGTVVVGIVILIVTGIKGRKK KNETKREENPYDSMDIGKGESNAGFQNSDDAQTSF
>NP_001116542.1 angiotensin-converting enzyme 2 precursor [Sus scrofa] MSGSFWLLLSLIPVTAAQSTTEELAKTFLEKFNLEAEDLAYQSSLASWTINTNITDENIQKMNDARAKWS AFYEEQSRIAKTYPLDEIQTLILKRQLQALQQSGTSGLSADKSKRLNTILNTMSTIYSSGKVLDPNNPQE CLVLEPGLDEIMENSKDYSRRLWAWESWRAEVGKQLRPLYEEYVVLENEMARANNYEDYGDYWRGDYEVT GTGDYDYSRNQLMEDVERTFAEIKPLYEHLHAYVRAKLMDAYPSRISPTGCLPAHLLGDMWGRFWTNLYP LTVPFGEKPSIDVTEAMVNQSWDAIRIFEEAEKFFVSIGLPNMTQGFWNNSMLTEPGDGRKVVCHPTAWD LGKGDFRIKMCTKVTMDDFLTAHHEMGHIQYDMAYAIQPYLLRNGANEGFHEAVGEIMSLSAATPHYLKA LGLLPPDFYEDSETEINFLLKQALTIVGTLPFTYMLEKWRWMVFKGEIPKEQWMQKWWEMKREIVGVVEP LPHDETYCDPACLFHVAEDYSFIRYYTRTIYQFQFHEALCRTAKHEGPLYKCDISNSTEAGQKLLQMLSL GKSEPWTLALENIVGVKTMDVKPLLSYFEPLLTWLKAQNGNSSVGWNTDWTPYADQSIKVRISLKSALGE DAYEWNDNEMYLFRSSIAYAMRNYFSSAKNETIPFGAVDVWVSDLKPRISFNFFVTSPANMSDIIPRSDV EKAISMSRSRINDAFRLDDNTLEFLGIQPTLGPPDEPPVTVWLIIFGVVMGLVVVGIVVLIFTGIRDRRK KKQASSEENPYGSMDLSKGESNSGFQNGDDIQTSF
>BAE53380.1 angiotensin I converting enzyme 2 [Mustela putorius furo] MLGSSWLLLSLAALTAAQSTTEDLAKTFLEKFNYEAEELSYQNSLASWNYNTNITDENIQKMNIAGAKWS AFYEEESQHAKTYPLEEIQDPIIKRQLRALQQSGSSVLSADKRERLNTILNAMSTIYSTGKACNPNNPQE CLLLEPGLDDIMENSKDYNERLWAWEGWRSEVGKQLRPLYEEYVALKNEMARANNYEDYGDYWRGDYEEE WADGYSYSRNQLIEDVEHTFTQIKPLYEHLHAYVRAKLMDAYPSRISPTGCLPAHLLGDMWGRFWTNLYP LMVPFRQKPNIDVTDAMVNQSWDARRIFEEAETFFVSVGLPNMTEGFWQNSMLTEPGDNRKVVCHPTAWD LGKRDFRIKMCTKVTMDDFLTAHHEMGHIQYDMAYAEQPFLLRNGANEGFHEAVGEIMSLSAATPNHLKN IGLLPPDFSEDSETDINFLLKQALTIVGTLPFTYMLEKWRWMVFKGEIPKEQWMQKWWEMKRDIVGVVEP LPHDETYCDPAALFHVANDYSFIRYYTRTIYQFQFQEALCQIAKHEGPLYKCDISNSSEAGQKLHEMLSL GRSKPWTFALERVVGAKTMDVRPLLNYFEPLFTWLKEQNRNSFVGWNTDWSPYADQSIKVRISLKSALGE KAYEWNDNEMYFFQSSIAYAMREYFSKVKNQTIPFVGKDVRVSDLKPRISFNFIVTSPENMSDIIPRADV EEAIRKSRGRINDAFRLDDNSLEFLGIQPTLEPPYQPPVTIWLIVFGVVMGVVVVGIFLLIFSGIRNRRK NNQARSEENPYASVDLSKGENNPGFQNVDDVQTSF
>AAX59005.1 angiotensin I converting enzyme 2 [Felis catus] MSGSFWLLLSFAALTAAQSTTEELAKTFLEKFNHEAEELSYQSSLASWNYNTNITDENVQKMNEAGAKWS AFYEEQSKLAKTYPLAEIHNTTVKRQLQALQQSGSSVLSADKSQRLNTILNAMSTIYSTGKACNPNNPQE CLLLEPGLDDIMENSKDYNERLWAWEGWRAEVGKQLRPLYEEYVALKNEMARANNYEDYGDYWRGDYEEE WTDGYNYSRSQLIKDVEHTFTQIKPLYQHLHAYVRAKLMDTYPSRISPTGCLPAHLLGDMWGRFWTNLYP LTVPFGQKPNIDVTDAMVNQSWDARRIFKEAEKFFVSVGLPNMTQGFWENSMLTEPGDSRKVVCHPTAWD LGKGDFRIKMCTKVTMDDFLTAHHEMGHIQYDMAYAVQPFLLRNGANEGFHEAVGEIMSLSAATPNHLKT IGLLSPGFSEDSETEINFLLKQALTIVGTLPFTYMLEKWRWMVFKGEIPKEQWMQKWWEMKREIVGVVEP VPHDETYCDPASLFHVANDYSFIRYYTRTIYQFQFQEALCRIAKHEGPLHKCDISNSSEAGKKLLQMLTL GKSKPWTLALEHVVGEKKMNVTPLLKYFEPLFTWLKEQNRNSFVGWNTDWRPYADQSIKVRISLKSALGD EAYEWNDNEMYLFRSSVAYAMREYFSKVKNQTIPFVEDNVWVSNLKPRISFNFFVTASKNVSDVIPRSEV EEAIRMSRSRINDAFRLDDNSLEFLGIQPTLSPPYQPPVTIWLIVFGVVMGVVVVGIVLLIVSGIRNRRK NNQARSEENPYASVDLSKGENNPGFQHADDVQTSF
>NP_001124604.1 angiotensin-converting enzyme 2 precursor [Pongo abelii] MSGSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLASWNYNTNITEENVQNMNNAGDKWS AFLKEQSTLAQMYPLQEIQNLTVKLQLQALQQNGSSVLSEDKSKRLNTILNTMSTIYSTGKVCNPNNPQE CLLLEPGLNEIMANSLDYNERLWAWESWRSEVGKQLRPLYEEYVVLKNEMARANHYEDYGDYWRGDYEVN GVDSYDYSRGQLIEDVEHTFEEIKPLYEHLHAYVRAKLINAYPSYISPIGCLPAHLLGDMWGRFWTNLYS LTVPFGQKPNIDVTDAMVDQAWDAQRIFKEAEKFFVSVGLPNMTQRFWENSMLTDPGNVQKVVCHPTAWD LGKGDFRILMCTKVTMDDFLTAHHEMGHIQYDMAYAAQPFLLRNGANEGFHEAVGEIMSLSAATPKHLKS IGLLSPDFQEDNETEINFLLKQALTIVGTLPFTYMLEKWRWMVFKGEIPKDQWMKKWWEMKREIVGVVEP VPHDETYCDPASLFHVSNDYSFIRYYTRTLYQFQFQEALCQAAKHEGPLHKCDISNSTEAGQKLLNMLRL GKSEPWTLALENVVGAKNMNVRPLLDYFEPLFTWLKDQNKNSFVGWSTDWSPYADQSIKVRISLKSALGN KAYEWNDNEIYLFRSSVAYAMRKYFLEVKNQMILFGEEDVRVANLKPRISFNFFVTAPKNVSDIIPRTEV EKAIRMSRSRINDAFRLNDNSLEFLGIQPTLGPPNQPPVSIWLIVFGVVMGVIVVGIVVLIFTGIRDRKK KNKARNEENPYASIDISKGENNPGFQNTDDVQTSF >AAY57872.1 angiotensin converting enzyme 2 [Chlorocebus aethiops] MSSSSWLLLSLVAVTAAQSTIEEQAKTFLDKFNHEAEDLFYQSSLASWNYNTNITEENVQNMNNAGEKWS AFLKEQSTLAQMYPLQAIQNLTVKLQLQALQQNGSSVLSEDKSKRLNTILNTMSTIHSTGKVCNPNNPQE CLLLDPGLNEIMEKSLDYNERLWAWEGWRSEVGKQLRPLYEEYVVLKNEMARANHYKDYGDYWRGDYEVN GVDGYDYNRDQLIEDVERTFEEIKPLYEHLHAYVRAKLMNAYPSYISPTGCLPAHLLGDMWGRFWTNLYS LTVPFGQKPNIDVTDAMVNQAWNAQRIFKEAEKFFVSVGLPNMTQGFWENSMLTDPGNVQKVVCHPTAWD LGKGDFRIIMCTKVTMDDFLTAHHEMGHIQYDMAYAAQPFLLRNGANEGFHEAVGEIMSLSAATPKHLKS IGLLSPDFQEDNETEINFLLKQALTIVGTLPFTYMLEKWRWMVFKGEIPKDQWMKKWWEMKREIVGVVEP VPHDETYCDPASLFHVSNDYSFIRYYTRTLYQFQFQEALCQAAKHEGPLHKCDISNSTEAGQKLLNMLKL GKSEPWTLALENVVGAKNMSVRPLLNYFEPLFTWLKDQNKNSFVGWSTDWSPYADQSIKVRISLKSALGA NAYKWNDNEMYLFRSSVAYAMRQYFLENKHQTILFGEEDVRVADLKPRISFNFYVTAPKNVSDIIPRTEV EEAIRFSRSRINDAFQLNDNSLEFLGIQSTLVPPYQSPITTWLIVFGVVMAVIVAGIVVLIFTGIRDRKK KNQARSEENPYASIDISKGENNPGFQNTDDVQTSF
Part 2: Creating a sequence alignment with Phylogeny.fr
- I went to the website www.phylogeny.fr. Then, I clicked 'Phylogeny analysis’, and clicked on the text ‘One Click'.
- Then, I clicked on ‘Upload your set of sequences in FASTA, EMBL, or NEXUS format’. I copied the protein sequences from Week 4 Talk Page.
- I used Command-V to paste my sequences in the field and clicked 'Submit".
- In order to properly align the sequences, I first pasted them into a Word document.
- I found the numbered tabs located just beneath the text One Click Mode, and clicked on the tab labeled 3. Alignment. Prior to this, I saw the pages named Alignment results, Phylogeny results, and Tree rendering results.
- Positions are color-coded to indicate their conservation. Blue highlighting meant high conservation (the sequences are identical or very similar), gray highlighting means lower conservation, and white highlighting means little conservation.
- Under Outputs, I clicked on Alignment in Clustal Format.
- This showed my sequences with the amount of conservation indicated below them. The amount of conservation corresponded to the color-coded highlights shown above.
- Key:
- “*” for invariant
- “:” for highly conserved
- “.” for weakly conserved
- Space for not conserved
- Below are the class' alignments
XP_0061228 ----------MLSHL---------------WILC--SLTVVVKSQDITQE-AINFLSEFN
XP_416822. ----------MLLHF---------------WLLC--GLSAVVTPQDVTQE-AQTFLAEFN
ETE61880.1 MLMKQAPVRKPSSRSFTHPAFFDLKGNMLTWLCLTWSLVVLALAQDETK-VATKFLEQFD
AGZ48803.1 ----------MSGSS---------------WLLL--SLVAVTTAQSTTEDEAKMFLDKFN
NP_0011165 ----------MSGSF---------------WLLL--SLIPVTAAQSTTEELAKTFLEKFN
XP_0313017 ----------MSGSF---------------WLLL--SLVAVTAAQSTTEELAKTFLEEFN
QLH93383.1 ----------MSGSS---------------WLLL--SLVAVTAAQSTSDEEAKTFLEKFN
BAE53380.1 ----------MLGSS---------------WLLL--SLAALTAAQSTTEDLAKTFLEKFN
CCP86723.1 ------------------------------------------------------------
XP_0258425 ----------MSGSS---------------WLLL--SLAALTAAQST-EDLVNTFLEKFN
AAX63775.1 ----------MSGSF---------------WLLL--SFAALTAAQSTTEELAKTFLETFN
AAX59005.1 ----------MSGSF---------------WLLL--SFAALTAAQSTTEELAKTFLEKFN
NP_0011239 ----------MSSSS---------------WLLL--SLVAVTTAQSLTEENAKTFLNNFN
AAW78017.1 ----------MSSSC---------------WLLL--SLVAVATAQSLIEEKAESFLNKFN
XP_0053160 MGSCPGARGKMLGSS---------------WLLL--SFVAVTAAQSTIEELAKTFLDKFN
AAY57872.1 ----------MSSSS---------------WLLL--SLVAVTAAQSTIEEQAKTFLDKFN
NP_0013583 ----------MSSSS---------------WLLL--SLVAVTAAQSTIEEQAKTFLDKFN
NP_0011246 ----------MSGSS---------------WLLL--SLVAVTAAQSTIEEQAKTFLDKFN
XP_0061228 VQAEDLSYASSLASWNYNTNITDENAKKMNEAGAKWSVFYDEASTNASKYAIDKITNHTV
XP_416822. VRAEDISYENSLASWNYNTNITEETARKMSEAGAKWAAFYEEASRNASRFSLANIQDAVT
ETE61880.1 ARATDLYYNASIASWNYNTNLTEENAKIMHEKDNIFSKFYGEACRNASMFNVNHITDETI
AGZ48803.1 TKAEDLSHQSSLASWDYNTNINDENVQKMDEAGAKWSAFYEEQSKLAKNYSLEQIQNVTV
NP_0011165 LEAEDLAYQSSLASWTINTNITDENIQKMNDARAKWSAFYEEQSRIAKTYPLDEIQTLIL
XP_0313017 HEAEDLSYQSSLASWNYNTNITDENVQKMNDARAKWSTFYEEKSKTAKTYPLEEIQNVTL
QLH93383.1 SEAEELSYQSSLASWNYNTNITDENVQKMNVAGAKWSTFYEEQSKIAKNYQLQNIQNDTI
BAE53380.1 YEAEELSYQNSLASWNYNTNITDENIQKMNIAGAKWSAFYEEESQHAKTYPLEEIQDPII
CCP86723.1 ------------------------------------------------------------
XP_0258425 YEAEELSYQSSLASWDYNTNISDENVQKMNNAGAKWSAFYEEQSKLAKTYPLEEIQDSTV
AAX63775.1 YEAQELSYQSSVASWNYNTNITDENAKNMNEAGAKWSAYYEEQSKLAQTYPLAEIQDAKI
AAX59005.1 HEAEELSYQSSLASWNYNTNITDENVQKMNEAGAKWSAFYEEQSKLAKTYPLAEIHNTTV
NP_0011239 QEAEDLSYQSSLASWNYNTNITEENAQKMSEAAAKWSAFYEEQSKTAQSFSLQEIQTPII
AAW78017.1 QEAEDLSYQSSLASWNYNTNITEENAQKMNEAAAKWSAFYEEQSKIAQNFSLQEIQNATI
XP_0053160 QEAEDLDYQRSLAAWNYNTNITEENTQKMNEAEAKWSAFYEEQSKLATAYPLQEIQNFTL
AAY57872.1 HEAEDLFYQSSLASWNYNTNITEENVQNMNNAGEKWSAFLKEQSTLAQMYPLQAIQNLTV
NP_0013583 HEAEDLFYQSSLASWNYNTNITEENVQNMNNAGDKWSAFLKEQSTLAQMYPLQEIQNLTV
NP_0011246 HEAEDLFYQSSLASWNYNTNITEENVQNMNNAGDKWSAFLKEQSTLAQMYPLQEIQNLTV
XP_0061228 KLQLQSLQGKGTSVLSGEKYNELNKILSTMSTFYSTGTVCKPDNPDICLPLEPGLDAIMA
XP_416822. RLQIQSLQDRGSSVLSPEKYSRLNSVMNSMSTIYSTGVVCKATEPFDCLVLEPGLDDIMA
ETE61880.1 KLQIRLLQ-SGSTDSTKD---QLDTVLHKMSTLYS-------------------LDDIMA
AGZ48803.1 KLQLQILQQSGSPVLSEDKSKRLNSILNAMSTIYSTGKVCKPNKPQECLLLEPGLDNIMG
NP_0011165 KRQLQALQQSGTSGLSADKSKRLNTILNTMSTIYSSGKVLDPNNPQECLVLEPGLDEIME
XP_0313017 KRQLQALQQSGASALSADKSKRLTTVLSTMSTIYSSGEVCDPNNPQECLVLEPGLDDIME
QLH93383.1 KRQLQALQLSGSSALSADKNQRLNTILNTMSTIYSTGKVCNPGNPQECSLLEPGLDNIME
BAE53380.1 KRQLRALQQSGSSVLSADKRERLNTILNAMSTIYSTGKACNPNNPQECLLLEPGLDDIME
CCP86723.1 ------------------------------------------------------------
XP_0258425 KRQLRALQHSGSSVLSADKNQRLNTILNSMSTIYSTGKACNPSNPQECLLLEPGLDDIME
AAX63775.1 KRQLQALQQSGSSVLSADKSQRLNTILNAMSTIYSTGKACNPNNPQECLLLEPGLDNIME
AAX59005.1 KRQLQALQQSGSSVLSADKSQRLNTILNAMSTIYSTGKACNPNNPQECLLLEPGLDDIME
NP_0011239 KRQLQALQQSGSSALSADKNKQLNTILNTMSTIYSTGKVCNPKNPQECLLLEPGLDEIMA
AAW78017.1 KRQLKALQQSGSSALSPDKNKQLNTILNTMSTIYSTGKVCNSMNPQECFLLEPGLDEIMA
XP_0053160 KRQLQALQQSGSSALSANKREQLNTILNTMSTIYSTGKVCNPKKPQECLLLEPGLDEIMA
AAY57872.1 KLQLQALQQNGSSVLSEDKSKRLNTILNTMSTIHSTGKVCNPNNPQECLLLDPGLNEIME
NP_0013583 KLQLQALQQNGSSVLSEDKSKRLNTILNTMSTIYSTGKVCNPDNPQECLLLEPGLNEIMA
NP_0011246 KLQLQALQQNGSSVLSEDKSKRLNTILNTMSTIYSTGKVCNPNNPQECLLLEPGLNEIMA
XP_0061228 SSTDYFERLWAWEGWRADVGKKMRELYERYVELENEAARLNKYSDYGDYWRGNYEVNDPT XP_416822. NSIDYHERLWAWEGWRADVGRMMRPLYEEYVELKNEAARLNNYSDYGDYWRANYETDYPE ETE61880.1 NNWNYPERLWAWEGWRANVGKKMRPLYETYVELKNKYARLRGYADYGDYWRANYEVDLPG AGZ48803.1 TSKDYNERLWAWEGWRAEVGKQLRPLYEEYVVLKNEMARGYHYEDYGDYWRRDYETEESP NP_0011165 NSKDYSRRLWAWESWRAEVGKQLRPLYEEYVVLENEMARANNYEDYGDYWRGDYEVTGTG XP_0313017 NSKDYNQRLWAWEGWRAEVGKQLRPLYEEYVVLKNEMARANNYEDYGDYWRGDYEVMWAG QLH93383.1 SSKDYNERLWAWEGWRSEVGKQLRPLYEEYVVLKNEMARANHYEDYGDYWRGDYETEGAN BAE53380.1 NSKDYNERLWAWEGWRSEVGKQLRPLYEEYVALKNEMARANNYEDYGDYWRGDYEEEWAD CCP86723.1 ------------------------------------------------------------ XP_0258425 NSKDYNERLWAWEGWRSEVGKQLRPLYEEYVALKNEMARANNYEDYGDYWRGDYEEEWEN AAX63775.1 NSKDYNERLWAWEGWRAEVGKQLRPLYEEYVALKNEMARANNYEDYGDYWRGDYEEEWTG AAX59005.1 NSKDYNERLWAWEGWRAEVGKQLRPLYEEYVALKNEMARANNYEDYGDYWRGDYEEEWTD NP_0011239 TSTDYNSRLWAWEGWRAEVGKQLRPLYEEYVVLKNEMARANNYNDYGDYWRGDYEAEGAD AAW78017.1 TSTDYNRRLWAWEGWRAEVGKQLRPLYEEYVVLKNEMARANNYEDYGDYWRGDYEAEGVE XP_0053160 NSTDYNERLWVWEGWRSEVGKQLRPLYEEYVVLKNEMARANNYEDYGDYWRGDYEAEGAD AAY57872.1 KSLDYNERLWAWEGWRSEVGKQLRPLYEEYVVLKNEMARANHYKDYGDYWRGDYEVNGVD NP_0013583 NSLDYNERLWAWESWRSEVGKQLRPLYEEYVVLKNEMARANHYEDYGDYWRGDYEVNGVD NP_0011246 NSLDYNERLWAWESWRSEVGKQLRPLYEEYVVLKNEMARANHYEDYGDYWRGDYEVNGVD
XP_0061228 EYAYSRNQLMEDVEATFEQIKPLYRELHAYVRYRLEKFYGSDHISSTGCLPAHLLGDMWG
XP_416822. EYKYSRDQLVQDVEKTFEQIKPLYQHLHAYVRHRLEQVYGSELINPTGCLPAHLLGDMWG
ETE61880.1 KFQYQREQLITDVESTFKQ------QLHAYVRHHLYKRYGPELINPEGAIPAHLLGDMWG
AGZ48803.1 GPGYSRDQLMKDVERIFTEIKPLYEHLHAYVRAKLMDTY-PFHISPTGCLPAHLLGDMWG
NP_0011165 DYDYSRNQLMEDVERTFAEIKPLYEHLHAYVRAKLMDAY-PSRISPTGCLPAHLLGDMWG
XP_0313017 DYDYSRDQLMGDVEHTFAEIKPLYEHLHAYVRAKLMDVY-PSHISPTGCLPAHLLGDMWG
QLH93383.1 GYNYSRDHLIEDVEHIFTQIKPLYEHLHAYVRAKLMDNY-PSHISPTGCLPAHLLGDMWG
BAE53380.1 GYSYSRNQLIEDVEHTFTQIKPLYEHLHAYVRAKLMDAY-PSRISPTGCLPAHLLGDMWG
CCP86723.1 ------------------------------------------------------------
XP_0258425 GYNYSRNQLIDDVEHTFTQIMPLYQHLHAYVRTKLMDTY-PSYISPTGCLPAHLLGDMWG
AAX63775.1 GYNYSRNQLIQDVEDTFEQIKPLYQHLHAYVRAKLMDTY-PSRISRTGCLPAHLLGDMWG
AAX59005.1 GYNYSRSQLIKDVEHTFTQIKPLYQHLHAYVRAKLMDTY-PSRISPTGCLPAHLLGDMWG
NP_0011239 GYNYNRNQLIEDVERTFAEIKPLYEHLHAYVRRKLMDTY-PSYISPTGCLPAHLLGDMWG
AAW78017.1 GYNYNRNQLIEDVENTFKEIKPLYEQLHAYVRTKLMEVY-PSYISPTGCLPAHLLGDMWG
XP_0053160 GYGYNRNQLIEDVERTFAEIKPLYEHLHAYVRAKLMNTY-PSYISPTGCLPAHLLGDMWG
AAY57872.1 GYDYNRDQLIEDVERTFEEIKPLYEHLHAYVRAKLMNAY-PSYISPTGCLPAHLLGDMWG
NP_0013583 GYDYSRGQLIEDVEHTFEEIKPLYEHLHAYVRAKLMNAY-PSYISPIGCLPAHLLGDMWG
NP_0011246 SYDYSRGQLIEDVEHTFEEIKPLYEHLHAYVRAKLINAY-PSYISPIGCLPAHLLGDMWG
XP_0061228 RFWTNLYALTVPYPDKPNIDVTSEMVKKNWNATKIFKAAEDFFMSVGLYKMTEGFWKNSM
XP_416822. RFWTNLYNLTVPYPEKPNIDVTSAMAQKNWDAMKIFKTAEAFFASIGLYNMTEGFWTNSM
ETE61880.1 RFWTNLYPLMVPYPNKTSIDVTSAMEKKKWTVNSIFKAAEHFFISIGLFNMTVGFWKNSM
AGZ48803.1 RFWTNLYPLTVPFGQKPNIDVTDEMLKQGWDADRIFKEAEKFFVSVGLPNMTEGFWNNSM
NP_0011165 RFWTNLYPLTVPFGEKPSIDVTEAMVNQSWDAIRIFEEAEKFFVSIGLPNMTQGFWNNSM
XP_0313017 RFWTNLYSLTVPFGQKPNIDVTEAMENQSWDAKRIFKEAEKFFVSIGLPNMTQGFWDNSM
QLH93383.1 RFWTNLYPLTVPFRQKPNIDVTDAMVNQTWDANRIFKEAEKFFVSVGLPKMTQTFWENSM
BAE53380.1 RFWTNLYPLMVPFRQKPNIDVTDAMVNQSWDARRIFEEAETFFVSVGLPNMTEGFWQNSM
CCP86723.1 ----------------------------------------------GLPNMTEGFWQNSM
XP_0258425 RFWTNLYPLTVPFGQKPNIDVTNAMVNQSWDARKIFKEAEKFFVSVGLPNMTQGFWENSM
AAX63775.1 RFWTNLYPLTVPFGQKPNIDVTDAMVNQNWDARRIFKEAEKFFVSVGLPNMTQGFWENSM
AAX59005.1 RFWTNLYPLTVPFGQKPNIDVTDAMVNQSWDARRIFKEAEKFFVSVGLPNMTQGFWENSM
NP_0011239 RFWTNLYPLTVPFAQKPNIDVTDAMMNQGWDAERIFQEAEKFFVSVGLPHMTQGFWANSM
AAW78017.1 RFWTNLYPLTTPFLQKPNIDVTDAMVNQSWDAERIFKEAEKFFVSVGLPQMTPGFWTNSM
XP_0053160 RFWTNLYSLTVPFPEKPNIDVTDAMINQNWNAVRIFKEAEKFFVSVGLPNMTQGFWENSM
AAY57872.1 RFWTNLYSLTVPFGQKPNIDVTDAMVNQAWNAQRIFKEAEKFFVSVGLPNMTQGFWENSM
NP_0013583 RFWTNLYSLTVPFGQKPNIDVTDAMVDQAWDAQRIFKEAEKFFVSVGLPNMTQGFWENSM
NP_0011246 RFWTNLYSLTVPFGQKPNIDVTDAMVDQAWDAQRIFKEAEKFFVSVGLPNMTQRFWENSM
** :** ** ***
XP_0061228 ITEPNDGRKVVCHPTAWDMGKKDYRIKMCTKVSMDDFLTVHHEMGHIEYDMAYSNLSYLL
XP_416822. LTEPTDNRKVVCHPTAWDMGKNDYRIKMCTKVTMDDFLTAHHEMGHIEYDMAYSVQPFLL
ETE61880.1 LEEPKGGRKVVCHPTAWDMGKEDYRIKMCTKINMEDFLTAHHEMGHIEYDMAYANQPFLL
AGZ48803.1 LTEPGDGRKVVCHPTAWDLGKGDFRIKMCTKVTMEDFLTAHHEMGHIQYDMAYASQPYLL
NP_0011165 LTEPGDGRKVVCHPTAWDLGKGDFRIKMCTKVTMDDFLTAHHEMGHIQYDMAYAIQPYLL
XP_0313017 LTEPGDGRKVVCHPTAWDLGKGDFRIKMCTKVTMDDFLTAHHEMGHIQYDMAYAIQPFLL
QLH93383.1 LTEPGDGRKVVCHPTAWDLGKHDFRIKMCTKVTMDDFLTAHHEMGHIQYDMAYAMQPYLL
BAE53380.1 LTEPGDNRKVVCHPTAWDLGKRDFRIKMCTKVTMDDFLTAHHEMGHIQYDMAYAEQPFLL
CCP86723.1 LTEPGDNRKVVCHPTAWDLGKHDFRIKMCTKVTMDDFLTAHHEMGHIQYDMAYAAQPFLL
XP_0258425 LTEPSDSRKVVCHPTAWDLGKGDFRIKMCTKVTMDDFLTAHHEMGHIQYDMAYAAQPFLL
AAX63775.1 LTEPGDGRKVVCHPTAWDLGKGDFRIKMCTKVTMDDFLTAHHEMGHIQYDMAYAAQPFLL
AAX59005.1 LTEPGDSRKVVCHPTAWDLGKGDFRIKMCTKVTMDDFLTAHHEMGHIQYDMAYAVQPFLL
NP_0011239 LTEPADGRKVVCHPTAWDLGHGDFRIKMCTKVTMDNFLTAHHEMGHIQYDMAYARQPFLL
AAW78017.1 LTEPGDDRKVVCHPTAWDLGHGDFRIKMCTKVTMDNFLTAHHEMGHIQYDMAYAKQPFLL
XP_0053160 LTEPTDGRKVVCHPTAWDLQKGDFRIKMCTKVTMDNFLTAHHEMGHIQYDMAYAMQPYLL
AAY57872.1 LTDPGNVQKVVCHPTAWDLGKGDFRIIMCTKVTMDDFLTAHHEMGHIQYDMAYAAQPFLL
NP_0013583 LTDPGNVQKAVCHPTAWDLGKGDFRILMCTKVTMDDFLTAHHEMGHIQYDMAYAAQPFLL
NP_0011246 LTDPGNVQKVVCHPTAWDLGKGDFRILMCTKVTMDDFLTAHHEMGHIQYDMAYAAQPFLL
: :* . .*.********: : *:** ****:.*::***.*******:*****: .:**
XP_0061228 RSGANEGFHEAVGEIMSLSAATPKHLKSLDLLEPTFQEDNETDINFLLKQALTIVGTMPF
XP_416822. RNGANEGFHEAVGEIMSLSAATPQHLKSLDLLEPTFQEDEETEINFLLKQALTIVGTMPF
ETE61880.1 RNGANEGFHEAVGEIMSLSAATPKYLKSLGLLEPTFQEDAETDINFLLKQALTIVGTMPF
AGZ48803.1 RNGANEGFHEAVGEVMSLSVATPKHLKTMGLLSPDFREDNETEINFLLKQALNIVGTLPF
NP_0011165 RNGANEGFHEAVGEIMSLSAATPHYLKALGLLPPDFYEDSETEINFLLKQALTIVGTLPF
XP_0313017 RNGANEGFHEAVGEIMSLSAATPHYLKALGLLPADFYEDSETEINFLLKQALTIVGTLPF
QLH93383.1 RNGANEGFHEAVGEIMSLSAATPKHLKNIGLLPPDFYEDNETEINFLLKQALTIVGTLPF
BAE53380.1 RNGANEGFHEAVGEIMSLSAATPNHLKNIGLLPPDFSEDSETDINFLLKQALTIVGTLPF
CCP86723.1 RNGANEGFHEAVGEIMSLSAATPNHLKNIGLLPPDFSEDSETDINFLLKQALTIVGTLPF
XP_0258425 RNGANEGFHEAVGEIMSLSAATPNHLKNIGLLPPSFFEDSETEINFLLKQALTIVGTLPF
AAX63775.1 RNGANEGFHEAVGEIMSLSAATPNHLKTIGLLSPAFSEDNETEINFLLKQALTIVGTLPF
AAX59005.1 RNGANEGFHEAVGEIMSLSAATPNHLKTIGLLSPGFSEDSETEINFLLKQALTIVGTLPF
NP_0011239 RNGANEGFHEAVGEIMSLSAATPKHLKSIGLLPSDFQEDSETEINFLLKQALTIVGTLPF
AAW78017.1 RNGANEGFHEAVGEIMSLSAATPKHLKSIGLLPSNFQEDNETEINFLLKQALTIVGTLPF
XP_0053160 RNGANEGFHEAVGEIMSLSASTPKHLKSIGLLPSDFREDSETEINFLLKQALTIVGTLPF
AAY57872.1 RNGANEGFHEAVGEIMSLSAATPKHLKSIGLLSPDFQEDNETEINFLLKQALTIVGTLPF
NP_0013583 RNGANEGFHEAVGEIMSLSAATPKHLKSIGLLSPDFQEDNETEINFLLKQALTIVGTLPF
NP_0011246 RNGANEGFHEAVGEIMSLSAATPKHLKSIGLLSPDFQEDNETEINFLLKQALTIVGTLPF
*.************:****.:**::** :.** . * ** **:*********.****:**
XP_0061228 TYMLEKWRWMVFKGDIPKDEWMKKWWEMKRAIVGVVEPVPHDETYCDPAALFHVANDYSF
XP_416822. TYMLEKWRWMVFNGEITKQEWTKRWWKMKREIVGVVEPVPHDETYCDPAALFHVANDYSF
ETE61880.1 TYMLEKWRWMVFAEQIPKDQWMKKWWEMKREIVGVVEPLPHNEEYCDPAALFHVANDYSF
AGZ48803.1 TYMLEKWRWMVFKGEIPKEEWMKKWWEMKRKIVGVVEPVPHDETYCDPASLFHVANDYSF
NP_0011165 TYMLEKWRWMVFKGEIPKEQWMQKWWEMKREIVGVVEPLPHDETYCDPACLFHVAEDYSF
XP_0313017 TYMLEKWRWMVFKGEIPKEQWMQKWWEMKREIVGVVEPLPHDETYCDPACLFHVAEDYSF
QLH93383.1 TYMLEKWRWMVFSGQIPKEQWMKKWWEMKREIVGVVEPVPHDETYCDPASLFHVANDYSF
BAE53380.1 TYMLEKWRWMVFKGEIPKEQWMQKWWEMKRDIVGVVEPLPHDETYCDPAALFHVANDYSF
CCP86723.1 TYMLEKWRWMVFKGEIPKEQWMQKWWEMKRDIVGVVEPLPHDETYCDPAALFHVANDYSF
XP_0258425 TYMLEKWRWMVFKGEIPKDQWMKTWWEMKRNIVGVVEPVPHDETYCDPASLFHVANDYSF
AAX63775.1 TYMLEKWRWMVFKGAIPKEQWMQKWWEMKRNIVGVVEPVPHDETYCDPASLFHVANDYSF
AAX59005.1 TYMLEKWRWMVFKGEIPKEQWMQKWWEMKREIVGVVEPVPHDETYCDPASLFHVANDYSF
NP_0011239 TYMLEKWRWMVFRGEIPKEQWMKKWWEMKREIVGVVEPLPHDETYCDPASLFHVSNDYSF
AAW78017.1 TYMLEKWRWMVFQDKIPREQWTKKWWEMKREIVGVVEPLPHDETYCDPASLFHVSNDYSF
XP_0053160 TYMLEKWRWMVFKGEIPKDQWMKKWWEMKREIVGVMEPVPHDETYCDPAALYHVSNDFSF
AAY57872.1 TYMLEKWRWMVFKGEIPKDQWMKKWWEMKREIVGVVEPVPHDETYCDPASLFHVSNDYSF
NP_0013583 TYMLEKWRWMVFKGEIPKDQWMKKWWEMKREIVGVVEPVPHDETYCDPASLFHVSNDYSF
NP_0011246 TYMLEKWRWMVFKGEIPKDQWMKKWWEMKREIVGVVEPVPHDETYCDPASLFHVSNDYSF
************ *..::* : **:*** ****:**:**:* *****.*:**::*:**
XP_0061228 IRYYTRTIYQFQFQEALCKAANHGGLLHTCDITNSMAAGQKLRDMLALGRSQPWTKALES
XP_416822. IRYYTRTIYQFQFQEALCKAANHTGPLHKCDITNSTAAGGNLRQLLELGKSKPWTQALES
ETE61880.1 IRYYTRTIYQFQFQEALCKAAGHTKELYKCDISDSTNAGRILKDMLALGSSQPWTKALES
AGZ48803.1 IRYYTRTIFEFQFHEALCRIAQHDGPLHKCDISNSTDAGKKLHQMLSVGKSQAWTKTLED
NP_0011165 IRYYTRTIYQFQFHEALCRTAKHEGPLYKCDISNSTEAGQKLLQMLSLGKSEPWTLALEN
XP_0313017 IRYYTRTIYQFQFHEALCQIAKHEGPLYKCDISNSTEAGQKLLQMLSLGKSEPWTLALEG
QLH93383.1 IRYYTRTIYQFQFQEALCQTAKHEGPLHKCDISNSTEAGQKLLQMLSLGKSKPWTLALER
BAE53380.1 IRYYTRTIYQFQFQEALCQIAKHEGPLYKCDISNSSEAGQKLHEMLSLGRSKPWTFALER
CCP86723.1 IRYYTRTIYQFQFQEALCQIAKHEGPLYKCDISNSREAGQKLHEMLSLGRSKPWTFALER
XP_0258425 IRYYTRTIYQFQFQEALCQIAKHEGPLHKCDISNSSEAGQKLLEMLKLGKSKPWTYALEI
AAX63775.1 IRYYTRTIYQFQFQEALCQIAKHEGPLHKCDISNSTEAGKKLLEMLSLGRSEPWTLALER
AAX59005.1 IRYYTRTIYQFQFQEALCRIAKHEGPLHKCDISNSSEAGKKLLQMLTLGKSKPWTLALEH
NP_0011239 IRYYTRTIYQFQFQEALCQAAKYNGSLHKCDISNSTEAGQKLLKMLSLGNSEPWTKALEN
AAW78017.1 IRYYTRTIYQFQFQEALCQAAKHDGPLHKCDISNSTEAGQKLLNMLSLGNSGPWTLALEN
XP_0053160 IRYYTRTIYQFQFQEALCQAAKHEGPLHKCDISNSTEAGQKLLNMLRLGKSKPWTLALEN
AAY57872.1 IRYYTRTLYQFQFQEALCQAAKHEGPLHKCDISNSTEAGQKLLNMLKLGKSEPWTLALEN
NP_0013583 IRYYTRTLYQFQFQEALCQAAKHEGPLHKCDISNSTEAGQKLFNMLRLGKSEPWTLALEN
NP_0011246 IRYYTRTLYQFQFQEALCQAAKHEGPLHKCDISNSTEAGQKLLNMLRLGKSEPWTLALEN
*******:::***:****. * : *:.***::* ** * .:* :* * .** :**
XP_0061228 ITGEKKMNATPLLHYFEPLYQWLIKNNSGRAVGWNTFWSPYSGNAIKVRISLKTALGDNA
XP_416822. ATGEKYMNATPLLHYFEPLFNWLQKNNSGRSIGWNTDWTPYSDNAIKVRISLKAALGDDA
ETE61880.1 ITGSLKMDAKPFCQYFDPLLKWLEKTNSNENVGWNVNWTPYSKDAIKVRISLKAALGDDA
AGZ48803.1 IVDSRNMDVGPLLKYFEPLYTWLQEQNRKSYVGWNTDWSPYSDQSIKVRISLKSALGENA
NP_0011165 IVGVKTMDVKPLLSYFEPLLTWLKAQNGNSSVGWNTDWTPYADQSIKVRISLKSALGEDA
XP_0313017 LVGVKTMDVKPLLNYFEPLLTWLKDQNRNSFVGWSTDWTPYTDQSIKVRISLKSALGDKA
QLH93383.1 VVGTKNMDVRPLLNYFEPLLTWLKEQNKNSFVGWNTDWSPYAAQSIKVRISLKSALGEKA
BAE53380.1 VVGAKTMDVRPLLNYFEPLFTWLKEQNRNSFVGWNTDWSPYADQSIKVRISLKSALGEKA
CCP86723.1 VVGAKTMDVRPLLNYFEPLFTWLKEQNRNSFVGWNTDWSPYADQSIKVRISLKSALGEKA
XP_0258425 VVGAKNMDVRPLLNYFEPLFTWLKEQNRNSFVGWNTDWSPYADQSIKVRISLKSALGEKA
AAX63775.1 VVGAKNMNVTPLLNYFEPLFTWLKEQNRNSFVGWDTDWRPYSDQSIKVRISLKSALGEKA
AAX59005.1 VVGEKKMNVTPLLKYFEPLFTWLKEQNRNSFVGWNTDWRPYADQSIKVRISLKSALGDEA
NP_0011239 VVGARNMDVKPLLNYFQPLFDWLKEQNRNSFVGWNTEWSPYADQSIKVRISLKSALGANA
AAW78017.1 VVGSRNMDVKPLLNYFQPLFVWLKEQNRNSTVGWSTDWSPYADQSIKVRISLKSALGKNA
XP_0053160 VVGARNMDVRPLLNYFEPLFGWLKDQNRNSFVGWNTNWSPYTDQSIKVRISLKSALGEEA
AAY57872.1 VVGAKNMSVRPLLNYFEPLFTWLKDQNKNSFVGWSTDWSPYADQSIKVRISLKSALGANA
NP_0013583 VVGAKNMNVRPLLNYFEPLFTWLKDQNKNSFVGWSTDWSPYADQSIKVRISLKSALGDKA
NP_0011246 VVGAKNMNVRPLLDYFEPLFTWLKDQNKNSFVGWSTDWSPYADQSIKVRISLKSALGNKA
.. *.. *: **:** ** * :**.. * **: ::********:*** .*
XP_0061228 YEWDENELYFFKSSIAYAMRKYFLEVKNQTVSFQCTDIHVWAVTQRVSFYFAVSMPGNAT
XP_416822. YVWDASELFLFKSSIAYAMRKYFAKEKEQNVDFQVTDIHVGEETQRVSFYLTVSMPGNVS
ETE61880.1 YNWDESEMFLFKSTIAYAMQKYFLEVKNKTVPF---------------------------
AGZ48803.1 YEWNDNEMYLFRSSVAYAMREYFLKEKHQTILFGAENVWVSNLKPRISFNFHVTSPGNLS
NP_0011165 YEWNDNEMYLFRSSIAYAMRNYFSSAKNETIPFGAVDVWVSDLKPRISFNFFVTSPANMS
XP_0313017 YEWNDNEMYLFQSSLAYAMRKYFLKVQNQTILFGVEDVWVSDLKPRISFSFFVTSPKNVS
QLH93383.1 YEWNDSEMYLFRSSVAYAMREYFSKFKKQTIPFEEESVRVSDLKPRVSFIFFVTLPKNVS
BAE53380.1 YEWNDNEMYFFQSSIAYAMREYFSKVKNQTIPFVGKDVRVSDLKPRISFNFIVTSPENMS
CCP86723.1 YEWNDNEMYFFQSSIAYAMREYFSKVKKQTIPFVDKDVRVSDLKPRISFNFIVTSPENMS
XP_0258425 YEWNNNEMYLFRSSIAYAMRRYFSEVKKQTIPFVEDNVWVSDLKPRISFNFFVTSPGNVS
AAX63775.1 YEWNDNEMYLFRSSIAYAMREYFSKVKNQTIPFVEDNVWVSDLKPRISFNFFVTFSNNVS
AAX59005.1 YEWNDNEMYLFRSSVAYAMREYFSKVKNQTIPFVEDNVWVSNLKPRISFNFFVTASKNVS
NP_0011239 YEWTNNEMFLFRSSVAYAMRKYFSIIKNQTVPFLEEDVRVSDLKPRVSFYFFVTSPQNVS
AAW78017.1 YEWTDNEMYLFRSSVAYAMREYFSREKNQTVPFGEADVWVSDLKPRVSFNFFVTSPKNVS
XP_0053160 YQWNDNEMYLFRSSVAYAMRMYFSKVKNQTIPFGEKDVWVSDEKPRISFNFFVTAPQNVS
AAY57872.1 YKWNDNEMYLFRSSVAYAMRQYFLENKHQTILFGEEDVRVADLKPRISFNFYVTAPKNVS
NP_0013583 YEWNDNEMYLFRSSVAYAMRQYFLKVKNQMILFGEEDVRVANLKPRISFNFFVTAPKNVS
NP_0011246 YEWNDNEIYLFRSSVAYAMRKYFLEVKNQMILFGEEDVRVANLKPRISFNFFVTAPKNVS
* * .*:::*.*::****. ** : : : *
XP_0061228 DFIPKSEVETAIRMSRGRINEAFRLDDNTLEFEGLLPTLASPYEPPVTVWLILFGVVMGV
XP_416822. DIVPRADVEKAIRMSRGRISEAFRLDDNTLEFDGIVPTLATPYKPPVTIWLILFGVVMSL
ETE61880.1 ------------HLSRDRINEAFKLTDQTLEFIGLLPTLAPPYESPITVWLVAFGVVIGL
AGZ48803.1 DIIPRPEVEGAIRMSRSRINDAFRLDDNSLEFLGIQPTLGPPYQPPVTIWLIVFGVVMAV
NP_0011165 DIIPRSDVEKAISMSRSRINDAFRLDDNTLEFLGIQPTLGPPDEPPVTVWLIIFGVVMGL
XP_0313017 DIIPRTEVEEAIRMSRSRINDAFRLDDNSLEFLGIQPTLGPPYEPPVTVWLIIFGIVMGL
QLH93383.1 AVIPRAEVEEAIRMSRSRINDVFRLDDNSLEFLGIQPTLEPPYQPPVTIWLIVFGVVMGV
BAE53380.1 DIIPRADVEEAIRKSRGRINDAFRLDDNSLEFLGIQPTLEPPYQPPVTIWLIVFGVVMGV
CCP86723.1 DIIPRADVEEAIRKSRGRINDAFRLDDNSLEFLGIQPTLEPPYQPPVTIWLIVFGVVMGV
XP_0258425 DIIPRTEVEKAIRMYRGRINDVFRLDDNSLEFLGIQPTLGPSYEPPVTIWLIVFGVVMGV
AAX63775.1 DVIPRSEVEDAIRMSRSRINDAFRLDDNSLEFLGIEPTLSPPYRPPVTIWLIVFGVVMGA
AAX59005.1 DVIPRSEVEEAIRMSRSRINDAFRLDDNSLEFLGIQPTLSPPYQPPVTIWLIVFGVVMGV
NP_0011239 DVIPRSEVEDAIRMSRGRINDVFGLNDNSLEFLGIHPTLEPPYQPPVTIWLIIFGVVMAL
AAW78017.1 DIIPRSEVEEAIRMSRGRINDIFGLNDNSLEFLGIYPTLKPPYEPPVTIWLIIFGVVMGT
XP_0053160 DIIPRTDVEKAIRMSRGRINGVFRLDDNSLEFLGIQPTLGPPYQPPVTIWLIVFGVVMGL
AAY57872.1 DIIPRTEVEEAIRFSRSRINDAFQLNDNSLEFLGIQSTLVPPYQSPITTWLIVFGVVMAV
NP_0013583 DIIPRTEVEKAIRMSRSRINDAFRLNDNSLEFLGIQPTLGPPNQPPVSIWLIVFGVVMGV
NP_0011246 DIIPRTEVEKAIRMSRSRINDAFRLNDNSLEFLGIQPTLGPPNQPPVSIWLIVFGVVMGV
*.**. * * *::*** *: .** .. .*:: **: **:*:.
XP_0061228 IVVGVIVLIVTGQRDRRKRMKAGTNELVQTNAIDP------ELENGEVNPAFIKHEERQT
XP_416822. IVIGVIVLIITGQRDKRKKARGRANEAGSNCEVNPYD------EDGRSNKGFEQSEETQT
ETE61880.1 IVIGIITLEKAGSKN---------------------------------------------
AGZ48803.1 VVVGIVVLIITGIRDRRKTDQARSEE-------NPYS--SVDLSKGENNPGFQNGDDVQT
NP_0011165 VVVGIVVLIFTGIRDRRKKKQASSEE-------NPYG--SMDLSKGESNSGFQNGDDIQT
XP_0313017 VVVGIVVLIFTGIRDRRKKKQASTEE-------NPYG--SVDLSKGENNSGFQNGDDVQT
QLH93383.1 IVVGIVVLIFTGIRDRKKKNQARSEQ-------NPYA--SVDLSKGENNPGFQNVDDVQT
BAE53380.1 VVVGIFLLIFSGIRNRRKNNQARSEE-------NPYA--SVDLSKGENNPGFQNVDDVQT
CCP86723.1 VVVGIFLLIFSGIRNRRKNNQARSEE-------NPYA--SVDLSKG--------------
XP_0258425 VVVGIVLLIFSGIRNRRKNDQARGEE-------NPYA--SVDLSKGENNPGFQNVDDAQT
AAX63775.1 IVVGIVLLIVSGIRNRRKNDQAGSEE-------NPYA--SVDLNKGENNPGFQHADDVQT
AAX59005.1 VVVGIVLLIVSGIRNRRKNNQARSEE-------NPYA--SVDLSKGENNPGFQHADDVQT
NP_0011239 VVVGIIILIVTGIKGRKKKNETKREE-------NPYD--SMDIGKGESNAGFQNSDDAQT
AAW78017.1 VVVGIVILIVTGIKGRKKKNETKREE-------NPYD--SMDIGKGESNAGFQNSDDAQT
XP_0053160 IVVGIVILIFTGIRDRRRKNQTKREE-------NPYAESSMEMGKGENNPGYQNNDDVQT
AAY57872.1 IVAGIVVLIFTGIRDRKKKNQARSEE-------NPYA--SIDISKGENNPGFQNTDDVQT
NP_0013583 IVVGIVILIFTGIRDRKKKNKARSGE-------NPYA--SIDISKGENNPGFQNTDDVQT
NP_0011246 IVVGIVVLIFTGIRDRKKKNKARNEE-------NPYA--SIDISKGENNPGFQNTDDVQT
:* *:. * :* ..
XP_0061228 SF XP_416822. SF ETE61880.1 -- AGZ48803.1 SF NP_0011165 SF XP_0313017 SF QLH93383.1 SF BAE53380.1 SF CCP86723.1 -- XP_0258425 SF AAX63775.1 SF AAX59005.1 SF NP_0011239 SF AAW78017.1 SF XP_0053160 SF AAY57872.1 SF NP_0013583 SF NP_0011246 SF
New alignment:
AGZ48803.1 ----------MSGSS---------------WLLL--SLVAVTTAQSTTEDEAKMFLDKFN
NP_0011165 ----------MSGSF---------------WLLL--SLIPVTAAQSTTEELAKTFLEKFN
XP_0313017 ----------MSGSF---------------WLLL--SLVAVTAAQSTTEELAKTFLEEFN
QLH93383.1 ----------MSGSS---------------WLLL--SLVAVTAAQSTSDEEAKTFLEKFN
BAE53380.1 ----------MLGSS---------------WLLL--SLAALTAAQSTTEDLAKTFLEKFN
XP_0258425 ----------MSGSS---------------WLLL--SLAALTAAQST-EDLVNTFLEKFN
AAX63775.1 ----------MSGSF---------------WLLL--SFAALTAAQSTTEELAKTFLETFN
AAX59005.1 ----------MSGSF---------------WLLL--SFAALTAAQSTTEELAKTFLEKFN
NP_0011239 ----------MSSSS---------------WLLL--SLVAVTTAQSLTEENAKTFLNNFN
AAW78017.1 ----------MSSSC---------------WLLL--SLVAVATAQSLIEEKAESFLNKFN
XP_0053160 MGSCPGARGKMLGSS---------------WLLL--SFVAVTAAQSTIEELAKTFLDKFN
AAY57872.1 ----------MSSSS---------------WLLL--SLVAVTAAQSTIEEQAKTFLDKFN
NP_0013583 ----------MSSSS---------------WLLL--SLVAVTAAQSTIEEQAKTFLDKFN
NP_0011246 ----------MSGSS---------------WLLL--SLVAVTAAQSTIEEQAKTFLDKFN
ETE61880.1 MLMKQAPVRKPSSRSFTHPAFFDLKGNMLTWLCLTWSLVVLALAQDETKVATK-FLEQFD
XP_0061228 ----------MLSHL---------------WILC--SLTVVVKSQDITQEAIN-FLSEFN
XP_416822. ----------MLLHF---------------WLLC--GLSAVVTPQDVTQE-AQTFLAEFN
*: .: :. .*. . : ** *:
AGZ48803.1 TKAEDLSHQSSLASWDYNTNINDENVQKMDEAGAKWSAFYEEQSKLAKNYSLEQIQNVTV
NP_0011165 LEAEDLAYQSSLASWTINTNITDENIQKMNDARAKWSAFYEEQSRIAKTYPLDEIQTLIL
XP_0313017 HEAEDLSYQSSLASWNYNTNITDENVQKMNDARAKWSTFYEEKSKTAKTYPLEEIQNVTL
QLH93383.1 SEAEELSYQSSLASWNYNTNITDENVQKMNVAGAKWSTFYEEQSKIAKNYQLQNIQNDTI
BAE53380.1 YEAEELSYQNSLASWNYNTNITDENIQKMNIAGAKWSAFYEEESQHAKTYPLEEIQDPII
XP_0258425 YEAEELSYQSSLASWDYNTNISDENVQKMNNAGAKWSAFYEEQSKLAKTYPLEEIQDSTV
AAX63775.1 YEAQELSYQSSVASWNYNTNITDENAKNMNEAGAKWSAYYEEQSKLAQTYPLAEIQDAKI
AAX59005.1 HEAEELSYQSSLASWNYNTNITDENVQKMNEAGAKWSAFYEEQSKLAKTYPLAEIHNTTV
NP_0011239 QEAEDLSYQSSLASWNYNTNITEENAQKMSEAAAKWSAFYEEQSKTAQSFSLQEIQTPII
AAW78017.1 QEAEDLSYQSSLASWNYNTNITEENAQKMNEAAAKWSAFYEEQSKIAQNFSLQEIQNATI
XP_0053160 QEAEDLDYQRSLAAWNYNTNITEENTQKMNEAEAKWSAFYEEQSKLATAYPLQEIQNFTL
AAY57872.1 HEAEDLFYQSSLASWNYNTNITEENVQNMNNAGEKWSAFLKEQSTLAQMYPLQAIQNLTV
NP_0013583 HEAEDLFYQSSLASWNYNTNITEENVQNMNNAGDKWSAFLKEQSTLAQMYPLQEIQNLTV
NP_0011246 HEAEDLFYQSSLASWNYNTNITEENVQNMNNAGDKWSAFLKEQSTLAQMYPLQEIQNLTV
ETE61880.1 ARATDLYYNASIASWNYNTNLTEENAKIMHEKDNIFSKFYGEACRNASMFNVNHITDETI
XP_0061228 VQAEDLSYASSLASWNYNTNITDENAKKMNEAGAKWSVFYDEASTNASKYAIDKITNHTV
XP_416822. VRAEDISYENSLASWNYNTNITEETARKMSEAGAKWAAFYEEASRNASRFSLANIQDAVT
* :: : *:*:* ***:.:*. . * :: : * . * : : *
AGZ48803.1 KLQLQILQQSGSPVLSEDKSKRLNSILNAMSTIYSTGKVCKPNKPQECLLLEPGLDNIMG
NP_0011165 KRQLQALQQSGTSGLSADKSKRLNTILNTMSTIYSSGKVLDPNNPQECLVLEPGLDEIME
XP_0313017 KRQLQALQQSGASALSADKSKRLTTVLSTMSTIYSSGEVCDPNNPQECLVLEPGLDDIME
QLH93383.1 KRQLQALQLSGSSALSADKNQRLNTILNTMSTIYSTGKVCNPGNPQECSLLEPGLDNIME
BAE53380.1 KRQLRALQQSGSSVLSADKRERLNTILNAMSTIYSTGKACNPNNPQECLLLEPGLDDIME
XP_0258425 KRQLRALQHSGSSVLSADKNQRLNTILNSMSTIYSTGKACNPSNPQECLLLEPGLDDIME
AAX63775.1 KRQLQALQQSGSSVLSADKSQRLNTILNAMSTIYSTGKACNPNNPQECLLLEPGLDNIME
AAX59005.1 KRQLQALQQSGSSVLSADKSQRLNTILNAMSTIYSTGKACNPNNPQECLLLEPGLDDIME
NP_0011239 KRQLQALQQSGSSALSADKNKQLNTILNTMSTIYSTGKVCNPKNPQECLLLEPGLDEIMA
AAW78017.1 KRQLKALQQSGSSALSPDKNKQLNTILNTMSTIYSTGKVCNSMNPQECFLLEPGLDEIMA
XP_0053160 KRQLQALQQSGSSALSANKREQLNTILNTMSTIYSTGKVCNPKKPQECLLLEPGLDEIMA
AAY57872.1 KLQLQALQQNGSSVLSEDKSKRLNTILNTMSTIHSTGKVCNPNNPQECLLLDPGLNEIME
NP_0013583 KLQLQALQQNGSSVLSEDKSKRLNTILNTMSTIYSTGKVCNPDNPQECLLLEPGLNEIMA
NP_0011246 KLQLQALQQNGSSVLSEDKSKRLNTILNTMSTIYSTGKVCNPNNPQECLLLEPGLNEIMA
ETE61880.1 KLQIRLLQ-SGSTDSTKD---QLDTVLHKMSTLYS-------------------LDDIMA
XP_0061228 KLQLQSLQGKGTSVLSGEKYNELNKILSTMSTFYSTGTVCKPDNPDICLPLEPGLDAIMA
XP_416822. RLQIQSLQDRGSSVLSPEKYSRLNSVMNSMSTIYSTGVVCKATEPFDCLVLEPGLDDIMA
. *:. ** *:. : : * .:: ***::* *: **
AGZ48803.1 TSKDYNERLWAWEGWRAEVGKQLRPLYEEYVVLKNEMARGYHYEDYGDYWRRDYETEESP
NP_0011165 NSKDYSRRLWAWESWRAEVGKQLRPLYEEYVVLENEMARANNYEDYGDYWRGDYEVTGTG
XP_0313017 NSKDYNQRLWAWEGWRAEVGKQLRPLYEEYVVLKNEMARANNYEDYGDYWRGDYEVMWAG
QLH93383.1 SSKDYNERLWAWEGWRSEVGKQLRPLYEEYVVLKNEMARANHYEDYGDYWRGDYETEGAN
BAE53380.1 NSKDYNERLWAWEGWRSEVGKQLRPLYEEYVALKNEMARANNYEDYGDYWRGDYEEEWAD
XP_0258425 NSKDYNERLWAWEGWRSEVGKQLRPLYEEYVALKNEMARANNYEDYGDYWRGDYEEEWEN
AAX63775.1 NSKDYNERLWAWEGWRAEVGKQLRPLYEEYVALKNEMARANNYEDYGDYWRGDYEEEWTG
AAX59005.1 NSKDYNERLWAWEGWRAEVGKQLRPLYEEYVALKNEMARANNYEDYGDYWRGDYEEEWTD
NP_0011239 TSTDYNSRLWAWEGWRAEVGKQLRPLYEEYVVLKNEMARANNYNDYGDYWRGDYEAEGAD
AAW78017.1 TSTDYNRRLWAWEGWRAEVGKQLRPLYEEYVVLKNEMARANNYEDYGDYWRGDYEAEGVE
XP_0053160 NSTDYNERLWVWEGWRSEVGKQLRPLYEEYVVLKNEMARANNYEDYGDYWRGDYEAEGAD
AAY57872.1 KSLDYNERLWAWEGWRSEVGKQLRPLYEEYVVLKNEMARANHYKDYGDYWRGDYEVNGVD
NP_0013583 NSLDYNERLWAWESWRSEVGKQLRPLYEEYVVLKNEMARANHYEDYGDYWRGDYEVNGVD
NP_0011246 NSLDYNERLWAWESWRSEVGKQLRPLYEEYVVLKNEMARANHYEDYGDYWRGDYEVNGVD
ETE61880.1 NNWNYPERLWAWEGWRANVGKKMRPLYETYVELKNKYARLRGYADYGDYWRANYEVDLPG
XP_0061228 SSTDYFERLWAWEGWRADVGKKMRELYERYVELENEAARLNKYSDYGDYWRGNYEVNDPT
XP_416822. NSIDYHERLWAWEGWRADVGRMMRPLYEEYVELKNEAARLNNYSDYGDYWRANYETDYPE
.. :* ***.**.**::**. :* *** ** *:*: ** * ******* :**
AGZ48803.1 GPGYSRDQLMKDVERIFTEIKPLYEHLHAYVRAKLMDTY-PFHISPTGCLPAHLLGDMWG
NP_0011165 DYDYSRNQLMEDVERTFAEIKPLYEHLHAYVRAKLMDAY-PSRISPTGCLPAHLLGDMWG
XP_0313017 DYDYSRDQLMGDVEHTFAEIKPLYEHLHAYVRAKLMDVY-PSHISPTGCLPAHLLGDMWG
QLH93383.1 GYNYSRDHLIEDVEHIFTQIKPLYEHLHAYVRAKLMDNY-PSHISPTGCLPAHLLGDMWG
BAE53380.1 GYSYSRNQLIEDVEHTFTQIKPLYEHLHAYVRAKLMDAY-PSRISPTGCLPAHLLGDMWG
XP_0258425 GYNYSRNQLIDDVEHTFTQIMPLYQHLHAYVRTKLMDTY-PSYISPTGCLPAHLLGDMWG
AAX63775.1 GYNYSRNQLIQDVEDTFEQIKPLYQHLHAYVRAKLMDTY-PSRISRTGCLPAHLLGDMWG
AAX59005.1 GYNYSRSQLIKDVEHTFTQIKPLYQHLHAYVRAKLMDTY-PSRISPTGCLPAHLLGDMWG
NP_0011239 GYNYNRNQLIEDVERTFAEIKPLYEHLHAYVRRKLMDTY-PSYISPTGCLPAHLLGDMWG
AAW78017.1 GYNYNRNQLIEDVENTFKEIKPLYEQLHAYVRTKLMEVY-PSYISPTGCLPAHLLGDMWG
XP_0053160 GYGYNRNQLIEDVERTFAEIKPLYEHLHAYVRAKLMNTY-PSYISPTGCLPAHLLGDMWG
AAY57872.1 GYDYNRDQLIEDVERTFEEIKPLYEHLHAYVRAKLMNAY-PSYISPTGCLPAHLLGDMWG
NP_0013583 GYDYSRGQLIEDVEHTFEEIKPLYEHLHAYVRAKLMNAY-PSYISPIGCLPAHLLGDMWG
NP_0011246 SYDYSRGQLIEDVEHTFEEIKPLYEHLHAYVRAKLINAY-PSYISPIGCLPAHLLGDMWG
ETE61880.1 KFQYQREQLITDVESTFKQ------QLHAYVRHHLYKRYGPELINPEGAIPAHLLGDMWG
XP_0061228 EYAYSRNQLMEDVEATFEQIKPLYRELHAYVRYRLEKFYGSDHISSTGCLPAHLLGDMWG
XP_416822. EYKYSRDQLVQDVEKTFEQIKPLYQHLHAYVRHRLEQVYGSELINPTGCLPAHLLGDMWG
*.* :*: *** * : ****** .* . * . *. *.:**********
AGZ48803.1 RFWTNLYPLTVPFGQKPNIDVTDEMLKQGWDADRIFKEAEKFFVSVGLPNMTEGFWNNSM
NP_0011165 RFWTNLYPLTVPFGEKPSIDVTEAMVNQSWDAIRIFEEAEKFFVSIGLPNMTQGFWNNSM
XP_0313017 RFWTNLYSLTVPFGQKPNIDVTEAMENQSWDAKRIFKEAEKFFVSIGLPNMTQGFWDNSM
QLH93383.1 RFWTNLYPLTVPFRQKPNIDVTDAMVNQTWDANRIFKEAEKFFVSVGLPKMTQTFWENSM
BAE53380.1 RFWTNLYPLMVPFRQKPNIDVTDAMVNQSWDARRIFEEAETFFVSVGLPNMTEGFWQNSM
XP_0258425 RFWTNLYPLTVPFGQKPNIDVTNAMVNQSWDARKIFKEAEKFFVSVGLPNMTQGFWENSM
AAX63775.1 RFWTNLYPLTVPFGQKPNIDVTDAMVNQNWDARRIFKEAEKFFVSVGLPNMTQGFWENSM
AAX59005.1 RFWTNLYPLTVPFGQKPNIDVTDAMVNQSWDARRIFKEAEKFFVSVGLPNMTQGFWENSM
NP_0011239 RFWTNLYPLTVPFAQKPNIDVTDAMMNQGWDAERIFQEAEKFFVSVGLPHMTQGFWANSM
AAW78017.1 RFWTNLYPLTTPFLQKPNIDVTDAMVNQSWDAERIFKEAEKFFVSVGLPQMTPGFWTNSM
XP_0053160 RFWTNLYSLTVPFPEKPNIDVTDAMINQNWNAVRIFKEAEKFFVSVGLPNMTQGFWENSM
AAY57872.1 RFWTNLYSLTVPFGQKPNIDVTDAMVNQAWNAQRIFKEAEKFFVSVGLPNMTQGFWENSM
NP_0013583 RFWTNLYSLTVPFGQKPNIDVTDAMVDQAWDAQRIFKEAEKFFVSVGLPNMTQGFWENSM
NP_0011246 RFWTNLYSLTVPFGQKPNIDVTDAMVDQAWDAQRIFKEAEKFFVSVGLPNMTQRFWENSM
ETE61880.1 RFWTNLYPLMVPYPNKTSIDVTSAMEKKKWTVNSIFKAAEHFFISIGLFNMTVGFWKNSM
XP_0061228 RFWTNLYALTVPYPDKPNIDVTSEMVKKNWNATKIFKAAEDFFMSVGLYKMTEGFWKNSM
XP_416822. RFWTNLYNLTVPYPEKPNIDVTSAMAQKNWDAMKIFKTAEAFFASIGLYNMTEGFWTNSM
******* * .*: :*..****. * .: * . **: ** ** *:** :** ** ***
AGZ48803.1 LTEPGDGRKVVCHPTAWDLGKGDFRIKMCTKVTMEDFLTAHHEMGHIQYDMAYASQPYLL
NP_0011165 LTEPGDGRKVVCHPTAWDLGKGDFRIKMCTKVTMDDFLTAHHEMGHIQYDMAYAIQPYLL
XP_0313017 LTEPGDGRKVVCHPTAWDLGKGDFRIKMCTKVTMDDFLTAHHEMGHIQYDMAYAIQPFLL
QLH93383.1 LTEPGDGRKVVCHPTAWDLGKHDFRIKMCTKVTMDDFLTAHHEMGHIQYDMAYAMQPYLL
BAE53380.1 LTEPGDNRKVVCHPTAWDLGKRDFRIKMCTKVTMDDFLTAHHEMGHIQYDMAYAEQPFLL
XP_0258425 LTEPSDSRKVVCHPTAWDLGKGDFRIKMCTKVTMDDFLTAHHEMGHIQYDMAYAAQPFLL
AAX63775.1 LTEPGDGRKVVCHPTAWDLGKGDFRIKMCTKVTMDDFLTAHHEMGHIQYDMAYAAQPFLL
AAX59005.1 LTEPGDSRKVVCHPTAWDLGKGDFRIKMCTKVTMDDFLTAHHEMGHIQYDMAYAVQPFLL
NP_0011239 LTEPADGRKVVCHPTAWDLGHGDFRIKMCTKVTMDNFLTAHHEMGHIQYDMAYARQPFLL
AAW78017.1 LTEPGDDRKVVCHPTAWDLGHGDFRIKMCTKVTMDNFLTAHHEMGHIQYDMAYAKQPFLL
XP_0053160 LTEPTDGRKVVCHPTAWDLQKGDFRIKMCTKVTMDNFLTAHHEMGHIQYDMAYAMQPYLL
AAY57872.1 LTDPGNVQKVVCHPTAWDLGKGDFRIIMCTKVTMDDFLTAHHEMGHIQYDMAYAAQPFLL
NP_0013583 LTDPGNVQKAVCHPTAWDLGKGDFRILMCTKVTMDDFLTAHHEMGHIQYDMAYAAQPFLL
NP_0011246 LTDPGNVQKVVCHPTAWDLGKGDFRILMCTKVTMDDFLTAHHEMGHIQYDMAYAAQPFLL
ETE61880.1 LEEPKGGRKVVCHPTAWDMGKEDYRIKMCTKINMEDFLTAHHEMGHIEYDMAYANQPFLL
XP_0061228 ITEPNDGRKVVCHPTAWDMGKKDYRIKMCTKVSMDDFLTVHHEMGHIEYDMAYSNLSYLL
XP_416822. LTEPTDNRKVVCHPTAWDMGKNDYRIKMCTKVTMDDFLTAHHEMGHIEYDMAYSVQPFLL
: :* . .*.********: : *:** ****:.*::***.*******:*****: .:**
AGZ48803.1 RNGANEGFHEAVGEVMSLSVATPKHLKTMGLLSPDFREDNETEINFLLKQALNIVGTLPF
NP_0011165 RNGANEGFHEAVGEIMSLSAATPHYLKALGLLPPDFYEDSETEINFLLKQALTIVGTLPF
XP_0313017 RNGANEGFHEAVGEIMSLSAATPHYLKALGLLPADFYEDSETEINFLLKQALTIVGTLPF
QLH93383.1 RNGANEGFHEAVGEIMSLSAATPKHLKNIGLLPPDFYEDNETEINFLLKQALTIVGTLPF
BAE53380.1 RNGANEGFHEAVGEIMSLSAATPNHLKNIGLLPPDFSEDSETDINFLLKQALTIVGTLPF
XP_0258425 RNGANEGFHEAVGEIMSLSAATPNHLKNIGLLPPSFFEDSETEINFLLKQALTIVGTLPF
AAX63775.1 RNGANEGFHEAVGEIMSLSAATPNHLKTIGLLSPAFSEDNETEINFLLKQALTIVGTLPF
AAX59005.1 RNGANEGFHEAVGEIMSLSAATPNHLKTIGLLSPGFSEDSETEINFLLKQALTIVGTLPF
NP_0011239 RNGANEGFHEAVGEIMSLSAATPKHLKSIGLLPSDFQEDSETEINFLLKQALTIVGTLPF
AAW78017.1 RNGANEGFHEAVGEIMSLSAATPKHLKSIGLLPSNFQEDNETEINFLLKQALTIVGTLPF
XP_0053160 RNGANEGFHEAVGEIMSLSASTPKHLKSIGLLPSDFREDSETEINFLLKQALTIVGTLPF
AAY57872.1 RNGANEGFHEAVGEIMSLSAATPKHLKSIGLLSPDFQEDNETEINFLLKQALTIVGTLPF
NP_0013583 RNGANEGFHEAVGEIMSLSAATPKHLKSIGLLSPDFQEDNETEINFLLKQALTIVGTLPF
NP_0011246 RNGANEGFHEAVGEIMSLSAATPKHLKSIGLLSPDFQEDNETEINFLLKQALTIVGTLPF
ETE61880.1 RNGANEGFHEAVGEIMSLSAATPKYLKSLGLLEPTFQEDAETDINFLLKQALTIVGTMPF
XP_0061228 RSGANEGFHEAVGEIMSLSAATPKHLKSLDLLEPTFQEDNETDINFLLKQALTIVGTMPF
XP_416822. RNGANEGFHEAVGEIMSLSAATPQHLKSLDLLEPTFQEDEETEINFLLKQALTIVGTMPF
*.************:****.:**::** :.** . * ** **:*********.****:**
AGZ48803.1 TYMLEKWRWMVFKGEIPKEEWMKKWWEMKRKIVGVVEPVPHDETYCDPASLFHVANDYSF
NP_0011165 TYMLEKWRWMVFKGEIPKEQWMQKWWEMKREIVGVVEPLPHDETYCDPACLFHVAEDYSF
XP_0313017 TYMLEKWRWMVFKGEIPKEQWMQKWWEMKREIVGVVEPLPHDETYCDPACLFHVAEDYSF
QLH93383.1 TYMLEKWRWMVFSGQIPKEQWMKKWWEMKREIVGVVEPVPHDETYCDPASLFHVANDYSF
BAE53380.1 TYMLEKWRWMVFKGEIPKEQWMQKWWEMKRDIVGVVEPLPHDETYCDPAALFHVANDYSF
XP_0258425 TYMLEKWRWMVFKGEIPKDQWMKTWWEMKRNIVGVVEPVPHDETYCDPASLFHVANDYSF
AAX63775.1 TYMLEKWRWMVFKGAIPKEQWMQKWWEMKRNIVGVVEPVPHDETYCDPASLFHVANDYSF
AAX59005.1 TYMLEKWRWMVFKGEIPKEQWMQKWWEMKREIVGVVEPVPHDETYCDPASLFHVANDYSF
NP_0011239 TYMLEKWRWMVFRGEIPKEQWMKKWWEMKREIVGVVEPLPHDETYCDPASLFHVSNDYSF
AAW78017.1 TYMLEKWRWMVFQDKIPREQWTKKWWEMKREIVGVVEPLPHDETYCDPASLFHVSNDYSF
XP_0053160 TYMLEKWRWMVFKGEIPKDQWMKKWWEMKREIVGVMEPVPHDETYCDPAALYHVSNDFSF
AAY57872.1 TYMLEKWRWMVFKGEIPKDQWMKKWWEMKREIVGVVEPVPHDETYCDPASLFHVSNDYSF
NP_0013583 TYMLEKWRWMVFKGEIPKDQWMKKWWEMKREIVGVVEPVPHDETYCDPASLFHVSNDYSF
NP_0011246 TYMLEKWRWMVFKGEIPKDQWMKKWWEMKREIVGVVEPVPHDETYCDPASLFHVSNDYSF
ETE61880.1 TYMLEKWRWMVFAEQIPKDQWMKKWWEMKREIVGVVEPLPHNEEYCDPAALFHVANDYSF
XP_0061228 TYMLEKWRWMVFKGDIPKDEWMKKWWEMKRAIVGVVEPVPHDETYCDPAALFHVANDYSF
XP_416822. TYMLEKWRWMVFNGEITKQEWTKRWWKMKREIVGVVEPVPHDETYCDPAALFHVANDYSF
************ *..::* : **:*** ****:**:**:* *****.*:**::*:**
AGZ48803.1 IRYYTRTIFEFQFHEALCRIAQHDGPLHKCDISNSTDAGKKLHQMLSVGKSQAWTKTLED
NP_0011165 IRYYTRTIYQFQFHEALCRTAKHEGPLYKCDISNSTEAGQKLLQMLSLGKSEPWTLALEN
XP_0313017 IRYYTRTIYQFQFHEALCQIAKHEGPLYKCDISNSTEAGQKLLQMLSLGKSEPWTLALEG
QLH93383.1 IRYYTRTIYQFQFQEALCQTAKHEGPLHKCDISNSTEAGQKLLQMLSLGKSKPWTLALER
BAE53380.1 IRYYTRTIYQFQFQEALCQIAKHEGPLYKCDISNSSEAGQKLHEMLSLGRSKPWTFALER
XP_0258425 IRYYTRTIYQFQFQEALCQIAKHEGPLHKCDISNSSEAGQKLLEMLKLGKSKPWTYALEI
AAX63775.1 IRYYTRTIYQFQFQEALCQIAKHEGPLHKCDISNSTEAGKKLLEMLSLGRSEPWTLALER
AAX59005.1 IRYYTRTIYQFQFQEALCRIAKHEGPLHKCDISNSSEAGKKLLQMLTLGKSKPWTLALEH
NP_0011239 IRYYTRTIYQFQFQEALCQAAKYNGSLHKCDISNSTEAGQKLLKMLSLGNSEPWTKALEN
AAW78017.1 IRYYTRTIYQFQFQEALCQAAKHDGPLHKCDISNSTEAGQKLLNMLSLGNSGPWTLALEN
XP_0053160 IRYYTRTIYQFQFQEALCQAAKHEGPLHKCDISNSTEAGQKLLNMLRLGKSKPWTLALEN
AAY57872.1 IRYYTRTLYQFQFQEALCQAAKHEGPLHKCDISNSTEAGQKLLNMLKLGKSEPWTLALEN
NP_0013583 IRYYTRTLYQFQFQEALCQAAKHEGPLHKCDISNSTEAGQKLFNMLRLGKSEPWTLALEN
NP_0011246 IRYYTRTLYQFQFQEALCQAAKHEGPLHKCDISNSTEAGQKLLNMLRLGKSEPWTLALEN
ETE61880.1 IRYYTRTIYQFQFQEALCKAAGHTKELYKCDISDSTNAGRILKDMLALGSSQPWTKALES
XP_0061228 IRYYTRTIYQFQFQEALCKAANHGGLLHTCDITNSMAAGQKLRDMLALGRSQPWTKALES
XP_416822. IRYYTRTIYQFQFQEALCKAANHTGPLHKCDITNSTAAGGNLRQLLELGKSKPWTQALES
*******:::***:****. * : *:.***::* ** * .:* :* * .** :**
AGZ48803.1 IVDSRNMDVGPLLKYFEPLYTWLQEQNRKSYVGWNTDWSPYSDQSIKVRISLKSALGENA
NP_0011165 IVGVKTMDVKPLLSYFEPLLTWLKAQNGNSSVGWNTDWTPYADQSIKVRISLKSALGEDA
XP_0313017 LVGVKTMDVKPLLNYFEPLLTWLKDQNRNSFVGWSTDWTPYTDQSIKVRISLKSALGDKA
QLH93383.1 VVGTKNMDVRPLLNYFEPLLTWLKEQNKNSFVGWNTDWSPYAAQSIKVRISLKSALGEKA
BAE53380.1 VVGAKTMDVRPLLNYFEPLFTWLKEQNRNSFVGWNTDWSPYADQSIKVRISLKSALGEKA
XP_0258425 VVGAKNMDVRPLLNYFEPLFTWLKEQNRNSFVGWNTDWSPYADQSIKVRISLKSALGEKA
AAX63775.1 VVGAKNMNVTPLLNYFEPLFTWLKEQNRNSFVGWDTDWRPYSDQSIKVRISLKSALGEKA
AAX59005.1 VVGEKKMNVTPLLKYFEPLFTWLKEQNRNSFVGWNTDWRPYADQSIKVRISLKSALGDEA
NP_0011239 VVGARNMDVKPLLNYFQPLFDWLKEQNRNSFVGWNTEWSPYADQSIKVRISLKSALGANA
AAW78017.1 VVGSRNMDVKPLLNYFQPLFVWLKEQNRNSTVGWSTDWSPYADQSIKVRISLKSALGKNA
XP_0053160 VVGARNMDVRPLLNYFEPLFGWLKDQNRNSFVGWNTNWSPYTDQSIKVRISLKSALGEEA
AAY57872.1 VVGAKNMSVRPLLNYFEPLFTWLKDQNKNSFVGWSTDWSPYADQSIKVRISLKSALGANA
NP_0013583 VVGAKNMNVRPLLNYFEPLFTWLKDQNKNSFVGWSTDWSPYADQSIKVRISLKSALGDKA
NP_0011246 VVGAKNMNVRPLLDYFEPLFTWLKDQNKNSFVGWSTDWSPYADQSIKVRISLKSALGNKA
ETE61880.1 ITGSLKMDAKPFCQYFDPLLKWLEKTNSNENVGWNVNWTPYSKDAIKVRISLKAALGDDA
XP_0061228 ITGEKKMNATPLLHYFEPLYQWLIKNNSGRAVGWNTFWSPYSGNAIKVRISLKTALGDNA
XP_416822. ATGEKYMNATPLLHYFEPLFNWLQKNNSGRSIGWNTDWTPYSDNAIKVRISLKAALGDDA
.. *.. *: **:** ** * :**.. * **: ::********:*** .*
AGZ48803.1 YEWNDNEMYLFRSSVAYAMREYFLKEKHQTILFGAENVWVSNLKPRISFNFHVTSPGNLS
NP_0011165 YEWNDNEMYLFRSSIAYAMRNYFSSAKNETIPFGAVDVWVSDLKPRISFNFFVTSPANMS
XP_0313017 YEWNDNEMYLFQSSLAYAMRKYFLKVQNQTILFGVEDVWVSDLKPRISFSFFVTSPKNVS
QLH93383.1 YEWNDSEMYLFRSSVAYAMREYFSKFKKQTIPFEEESVRVSDLKPRVSFIFFVTLPKNVS
BAE53380.1 YEWNDNEMYFFQSSIAYAMREYFSKVKNQTIPFVGKDVRVSDLKPRISFNFIVTSPENMS
XP_0258425 YEWNNNEMYLFRSSIAYAMRRYFSEVKKQTIPFVEDNVWVSDLKPRISFNFFVTSPGNVS
AAX63775.1 YEWNDNEMYLFRSSIAYAMREYFSKVKNQTIPFVEDNVWVSDLKPRISFNFFVTFSNNVS
AAX59005.1 YEWNDNEMYLFRSSVAYAMREYFSKVKNQTIPFVEDNVWVSNLKPRISFNFFVTASKNVS
NP_0011239 YEWTNNEMFLFRSSVAYAMRKYFSIIKNQTVPFLEEDVRVSDLKPRVSFYFFVTSPQNVS
AAW78017.1 YEWTDNEMYLFRSSVAYAMREYFSREKNQTVPFGEADVWVSDLKPRVSFNFFVTSPKNVS
XP_0053160 YQWNDNEMYLFRSSVAYAMRMYFSKVKNQTIPFGEKDVWVSDEKPRISFNFFVTAPQNVS
AAY57872.1 YKWNDNEMYLFRSSVAYAMRQYFLENKHQTILFGEEDVRVADLKPRISFNFYVTAPKNVS
NP_0013583 YEWNDNEMYLFRSSVAYAMRQYFLKVKNQMILFGEEDVRVANLKPRISFNFFVTAPKNVS
NP_0011246 YEWNDNEIYLFRSSVAYAMRKYFLEVKNQMILFGEEDVRVANLKPRISFNFFVTAPKNVS
ETE61880.1 YNWDESEMFLFKSTIAYAMQKYFLEVKNKTVPF---------------------------
XP_0061228 YEWDENELYFFKSSIAYAMRKYFLEVKNQTVSFQCTDIHVWAVTQRVSFYFAVSMPGNAT
XP_416822. YVWDASELFLFKSSIAYAMRKYFAKEKEQNVDFQVTDIHVGEETQRVSFYLTVSMPGNVS
* * .*:::*.*::****. ** : : : *
AGZ48803.1 DIIPRPEVEGAIRMSRSRINDAFRLDDNSLEFLGIQPTLGPPYQPPVTIWLIVFGVVMAV
NP_0011165 DIIPRSDVEKAISMSRSRINDAFRLDDNTLEFLGIQPTLGPPDEPPVTVWLIIFGVVMGL
XP_0313017 DIIPRTEVEEAIRMSRSRINDAFRLDDNSLEFLGIQPTLGPPYEPPVTVWLIIFGIVMGL
QLH93383.1 AVIPRAEVEEAIRMSRSRINDVFRLDDNSLEFLGIQPTLEPPYQPPVTIWLIVFGVVMGV
BAE53380.1 DIIPRADVEEAIRKSRGRINDAFRLDDNSLEFLGIQPTLEPPYQPPVTIWLIVFGVVMGV
XP_0258425 DIIPRTEVEKAIRMYRGRINDVFRLDDNSLEFLGIQPTLGPSYEPPVTIWLIVFGVVMGV
AAX63775.1 DVIPRSEVEDAIRMSRSRINDAFRLDDNSLEFLGIEPTLSPPYRPPVTIWLIVFGVVMGA
AAX59005.1 DVIPRSEVEEAIRMSRSRINDAFRLDDNSLEFLGIQPTLSPPYQPPVTIWLIVFGVVMGV
NP_0011239 DVIPRSEVEDAIRMSRGRINDVFGLNDNSLEFLGIHPTLEPPYQPPVTIWLIIFGVVMAL
AAW78017.1 DIIPRSEVEEAIRMSRGRINDIFGLNDNSLEFLGIYPTLKPPYEPPVTIWLIIFGVVMGT
XP_0053160 DIIPRTDVEKAIRMSRGRINGVFRLDDNSLEFLGIQPTLGPPYQPPVTIWLIVFGVVMGL
AAY57872.1 DIIPRTEVEEAIRFSRSRINDAFQLNDNSLEFLGIQSTLVPPYQSPITTWLIVFGVVMAV
NP_0013583 DIIPRTEVEKAIRMSRSRINDAFRLNDNSLEFLGIQPTLGPPNQPPVSIWLIVFGVVMGV
NP_0011246 DIIPRTEVEKAIRMSRSRINDAFRLNDNSLEFLGIQPTLGPPNQPPVSIWLIVFGVVMGV
ETE61880.1 ------------HLSRDRINEAFKLTDQTLEFIGLLPTLAPPYESPITVWLVAFGVVIGL
XP_0061228 DFIPKSEVETAIRMSRGRINEAFRLDDNTLEFEGLLPTLASPYEPPVTVWLILFGVVMGV
XP_416822. DIVPRADVEKAIRMSRGRISEAFRLDDNTLEFDGIVPTLATPYKPPVTIWLILFGVVMSL
*.**. * * *::*** *: .** .. .*:: **: **:*:.
AGZ48803.1 VVVGIVVLIITGIRDRRKTDQARSEE-------NPYS--SVDLSKGENNPGFQNGDDVQT
NP_0011165 VVVGIVVLIFTGIRDRRKKKQASSEE-------NPYG--SMDLSKGESNSGFQNGDDIQT
XP_0313017 VVVGIVVLIFTGIRDRRKKKQASTEE-------NPYG--SVDLSKGENNSGFQNGDDVQT
QLH93383.1 IVVGIVVLIFTGIRDRKKKNQARSEQ-------NPYA--SVDLSKGENNPGFQNVDDVQT
BAE53380.1 VVVGIFLLIFSGIRNRRKNNQARSEE-------NPYA--SVDLSKGENNPGFQNVDDVQT
XP_0258425 VVVGIVLLIFSGIRNRRKNDQARGEE-------NPYA--SVDLSKGENNPGFQNVDDAQT
AAX63775.1 IVVGIVLLIVSGIRNRRKNDQAGSEE-------NPYA--SVDLNKGENNPGFQHADDVQT
AAX59005.1 VVVGIVLLIVSGIRNRRKNNQARSEE-------NPYA--SVDLSKGENNPGFQHADDVQT
NP_0011239 VVVGIIILIVTGIKGRKKKNETKREE-------NPYD--SMDIGKGESNAGFQNSDDAQT
AAW78017.1 VVVGIVILIVTGIKGRKKKNETKREE-------NPYD--SMDIGKGESNAGFQNSDDAQT
XP_0053160 IVVGIVILIFTGIRDRRRKNQTKREE-------NPYAESSMEMGKGENNPGYQNNDDVQT
AAY57872.1 IVAGIVVLIFTGIRDRKKKNQARSEE-------NPYA--SIDISKGENNPGFQNTDDVQT
NP_0013583 IVVGIVILIFTGIRDRKKKNKARSGE-------NPYA--SIDISKGENNPGFQNTDDVQT
NP_0011246 IVVGIVVLIFTGIRDRKKKNKARNEE-------NPYA--SIDISKGENNPGFQNTDDVQT
ETE61880.1 IVIGIITLEKAGSKN---------------------------------------------
XP_0061228 IVVGVIVLIVTGQRDRRKRMKAGTNELVQTNAIDP------ELENGEVNPAFIKHEERQT
XP_416822. IVIGVIVLIITGQRDKRKKARGRANEAGSNCEVNPYD------EDGRSNKGFEQSEETQT
:* *:. * :* ..
AGZ48803.1 SF NP_0011165 SF XP_0313017 SF QLH93383.1 SF BAE53380.1 SF XP_0258425 SF AAX63775.1 SF AAX59005.1 SF NP_0011239 SF AAW78017.1 SF XP_0053160 SF AAY57872.1 SF NP_0013583 SF NP_0011246 SF ETE61880.1 -- XP_0061228 SF XP_416822. SF
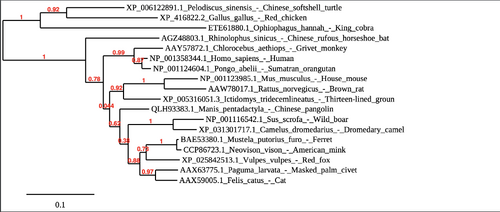
Part 3: Creating a phylogenetic tree with Phylogeny.fr
- Continued working with the www.phylogeny.fr website.
- Went back to 6. Tree rendering for the phylogenetic tree of the sequences.
- Horizontal lines represent individual evolutionary lines.
- Vertical lines represent mutation events. the vertical length has no biological meaning.
- The left-most split is called the root of the tree, which represents a hypothesis about the most recent common ancestor (MRCA) of the sequences within your tree.
- The length of each branch represents the percentage change in the amino acid sequence occurring along that branch, relative to the scale bar
- The scale bar was 0.5 (50%).
- I saved the image to a file and uploaded it to the wiki.
- Original image included the American mink. After further analysis of this partial sequence, this sequence was removed.
- The new phylogenetic tree is visible below:
Part 4: Structural Analysis and Critical Residues Table
- My research partner, Aiden Burnett, performed a structural analysis of the ACE2 receptor.
- Aiden Burnett also created a table detailing the differences between the critical amino acid residues.
- The methods by which he did these can both be found on his user page.
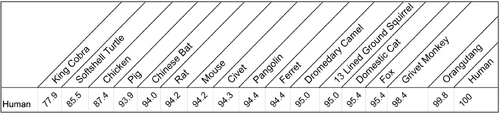
Part 5: Sequence Percent Similarity Table
- Navigated to LALIGN, a platform used by researchers to find a percent identity for sequences.
- Allows for the comparison of two different sequences.
- Entered human ACE2 sequence in area called 1st Query sequence.
- Entered one of the other seventeen sequences into the ;2nd Query sequence'.
- Clicked Run LALIGN.
- Noted the percent similarity between the sequences.
- Recorded this value in a table.
- Repeated procedure for other sixteen sequences, comparing each to the human ACE2 sequence.
- Created a table showing the percent similarity of each organism to humans.
Part 6: Final Presentation
- Uploaded presentation created based on the information presented above.
Scientific Conclusion
- Known human orthologues, including monkeys and orangutans, showed close similarities to the human ACE2 sequences. Foxes and cats also showed similarities, while turtles and king cobras did not show to be similar. Of the five critical amino acids that correspond to the RBD of SARS-CoV-2 on the ACE2 receptor, many were relatively conserved across species, with most organisms having between 2-3 of the 5 amino acids altered. This could help study the lineage of SARS-CoV-2 and identify which animals could act as intermediary hosts for future strains of SARS viruses.
Acknowledgments
- I consulted with my partner Aiden Burnett in class, over text, as well as over the phone several times to discuss the creation of our presentation.
- I contacted my TA, Annika Dinulos, to ask about the formatting of a presentation.
- I copied and modified procedures from the Week 6 assignment page.
- I referred back to procedures used on my Week 4 and Week 5 pages.
- I used the Wan et. al - Receptor Recognition by the Novel Coronavirus from Wuhan paper for reference.
- I obtained sequences from GenBank.
- I built the phylogenetic tree and created sequence alignments using Phylogeny.fr.
- I used LALIGN to compare sequence percent similarities.
- I uploaded images using the Wiki Upload page.
- I copied and modified wiki syntax on formatting a photo from the Media Wiki Help Page.
- Except for what is noted above, this individual journal entry was completed by me and not copied from another source.
Anna Horvath (talk) 21:25, 14 October 2020 (PDT)
References
- Andersen, K., Rambaut, A., Lipkin, W., Holmes, E., & Garry, R. (2020). The proximal origin of SARS-CoV-2. Nature Medicine, 26(4), 450-452. doi: 10.1038/s41591-020-0820-9
- Angiotensin-converting enzyme 2 isoform 1 precursor [Homo sapiens] - Protein - NCBI. (2020). Retrieved 8 October 2020, from https://www.ncbi.nlm.nih.gov/protein/NP_001358344.1?report=fasta
- Angiotensin-converting enzyme 2 [Paguma larvata] - Protein - NCBI. (2020). Retrieved 8 October 2020, from https://www.ncbi.nlm.nih.gov/protein/AAX63775.1?report=fasta
- Angiotensin-converting enzyme 2 [Rhinolophus sinicus] - Protein - NCBI. (2020). Retrieved 8 October 2020, from https://www.ncbi.nlm.nih.gov/protein/AGZ48803.1?report=fasta
- Angiotensin-converting enzyme 2 precursor [Mus musculus] - Protein - NCBI. (2020). Retrieved 8 October 2020, from https://www.ncbi.nlm.nih.gov/protein/NP_001123985.1?report=fasta
- angiotensin converting enzyme 2 [Rattus norvegicus] - Protein - NCBI. (2020). Retrieved 8 October 2020, from https://www.ncbi.nlm.nih.gov/protein/AAW78017.1?report=fasta
- Angiotensin-converting enzyme 2 precursor [Sus scrofa] - Protein - NCBI. (2020). Retrieved 8 October 2020, from https://www.ncbi.nlm.nih.gov/protein/NP_001116542.1?report=fasta
- Angiotensin I converting enzyme 2 [Mustela putorius furo] - Protein - NCBI. (2020). Retrieved 8 October 2020, from https://www.ncbi.nlm.nih.gov/protein/BAE53380.1?report=fasta
- Angiotensin I converting enzyme 2 [Felis catus] - Protein - NCBI. (2020). Retrieved 8 October 2020, from https://www.ncbi.nlm.nih.gov/protein/AAX59005.1?report=fasta
- Angiotensin-converting enzyme 2 precursor [Pongo abelii] - Protein - NCBI. (2020). Retrieved 8 October 2020, from https://www.ncbi.nlm.nih.gov/protein/NP_001124604.1?report=fasta
- Angiotensin converting enzyme 2 [Chlorocebus aethiops] - Protein - NCBI. (2020). Retrieved 8 October 2020, from https://www.ncbi.nlm.nih.gov/protein/AAY57872.1?report=fasta
- Angiotensin-converting enzyme 2 [Vulpes vulpes] - Protein - NCBI. (2020). Retrieved 8 October 2020, from https://www.ncbi.nlm.nih.gov/protein/XP_025842513.1?report=fasta
- Angiotensin-converting enzyme 2 [Gallus gallus] - Protein - NCBI. (2020). Retrieved 8 October 2020, from https://www.ncbi.nlm.nih.gov/protein/XP_416822.2?report=fasta
- Angiotensin-converting enzyme 2 [Ophiophagus hannah] - Protein - NCBI. (2020). Retrieved 8 October 2020, from https://www.ncbi.nlm.nih.gov/protein/ETE61880.1?report=fasta
- Angiotensin I converting enzyme 2 [Manis pentadactyla] - Protein - NCBI. (n.d.). Retrieved October 08, 2020, from https://www.ncbi.nlm.nih.gov/protein/QLH93383.1?report=fasta
- Angiotensin-converting enzyme 2 [Camelus dromedarius] - Protein - NCBI. (n.d.). Retrieved October 08, 2020, from https://www.ncbi.nlm.nih.gov/protein/XP_031301717.1
- Angiotensin-converting enzyme 2, partial [Neovison vison] - Protein - NCBI. (2020). Retrieved 8 October 2020, from https://www.ncbi.nlm.nih.gov/protein/CCP86723.1?report=fasta
- Angiotensin-converting enzyme 2 [Ictidomys tridecemlineatus] - Protein - NCBI. (2020). Retrieved 8 October 2020, from https://www.ncbi.nlm.nih.gov/protein/XP_005316051.3?report=fasta
- Angiotensin-converting enzyme 2 [Pelodiscus sinensis] - Protein - NCBI. (2020). Retrieved 8 October 2020, from https://www.ncbi.nlm.nih.gov/protein/XP_006122891.1?report=fasta
- Deng, J., Jin, Y., Liu, Y., Sun, J., Hao, L., & Bai, J. et al. (2020). Serological survey of SARS‐CoV‐2 for experimental, domestic, companion and wild animals excludes intermediate hosts of 35 different species of animals. Transboundary And Emerging Diseases, 67(4), 1745-1749. doi: 10.1111/tbed.13577
- LALIGN Server. (2020). Retrieved 14 October 2020, from https://embnet.vital-it.ch/software/LALIGN_form.html
- OpenWetWare - Anna Horvath Week 4. (2020). Retrieved 14 October 2020, from https://openwetware.org/wiki/Anna_Horvath_Week_4
- OpenWetWare - Anna Horvath Week 5. (2020). Retrieved 14 October 2020, from https://openwetware.org/wiki/Anna_Horvath_Week_5
- OpenWetWare - BIOL368/F20:Week 6. (2020). Retrieved 14 October 2020, from https://openwetware.org/wiki/BIOL368/F20:Week_6
- Phylogeny.fr: "One Click" Mode. (2020). Retrieved 8 October 2020, from http://www.phylogeny.fr/simple_phylogeny.cgi?workflow_id=b9c0813cbbe9695d63cf7e31da5f026d&tab_index=1
- Wan, Y., Shang, J., Graham, R., Baric, R., & Li, F. (2020). Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. Journal Of Virology, 94(7). doi: 10.1128/jvi.00127-20
- Yuan, S., Jiang, S., & Li, Z. (2020). Analysis of Possible Intermediate Hosts of the New Coronavirus SARS-CoV-2. Frontiers In Veterinary Science, 7. doi: 10.3389/fvets.2020.00379
- Zhao, J., Cui, W., & Tian, B. (2020). The Potential Intermediate Hosts for SARS-CoV-2. Frontiers In Microbiology, 11. doi: 10.3389/fmicb.2020.580137
Template
User Pages
Assignments
- Week 1 Assignment
- Week 2 Assignment
- Week 3 Assignment
- Week 4 Assignment
- Week 5 Assignment
- Week 6 Assignment
- Week 7 Assignment
- Week 8 Assignment
- Week 10 Assignment
- Week 11 Assignment
- Week 12 Assignment
- Week 14 Assignment
Journal Pages
- Anna Horvath Week 2
- Anna Horvath Week 3
- Anna Horvath Week 4
- Anna Horvath Week 5
- Anna Horvath Week 6
- Anna Horvath Week 7
- DrugComboDB Review
- Anna Horvath Week 10
- Anna Horvath Week 11
- Anna Horvath Week 12
- Anna Horvath Week 14