BioMicroCenter:DNA HTL: Difference between revisions
No edit summary |
|||
| Line 19: | Line 19: | ||
|INCLUDED || Library preparation <BR> Spot check of final libraries | |INCLUDED || Library preparation <BR> Spot check of final libraries | ||
|- | |- | ||
|SUBMISSION || MIT - [https://mit.ilabsolutions.com/service_item/new/3381?spt_id= | |SUBMISSION || MIT - [https://mit.ilabsolutions.com/service_item/new/3381?spt_id=3863 ilabs] <BR> External - [[BioMicroCenter:Forms|form]] | ||
|- | |- | ||
|UNIT || 24 samples <BR> 96 samples | |UNIT || 24 samples <BR> 96 samples | ||
| Line 50: | Line 50: | ||
|INCLUDED || Library preparation <BR> Spot check of final libraries | |INCLUDED || Library preparation <BR> Spot check of final libraries | ||
|- | |- | ||
|SUBMISSION || MIT - [https://mit.ilabsolutions.com/service_item/new/3381?spt_id= | |SUBMISSION || MIT - [https://mit.ilabsolutions.com/service_item/new/3381?spt_id=3863 ilabs] <BR> External - [[BioMicroCenter:Forms|form]] | ||
|- | |- | ||
|UNIT || 16 samples (full reaction)<BR> 96 samples <BR> 384 samples | |UNIT || 16 samples (full reaction)<BR> 96 samples <BR> 384 samples | ||
Revision as of 19:48, 22 October 2018
| HOME -- | SEQUENCING -- | LIBRARY PREP -- | HIGH-THROUGHPUT -- | COMPUTING -- | DATA MANAGEMENT -- | OTHER TECHNOLOGY |
The BioMicro Center offers high-throuhgput DNA library preparation for NexteraXT samples and for metagenomics/amplicon generation. High-throughput library generation is different from standard library preparation in a couple key ways. First, with dozens of samples to prepare, small numbers of samples may fail and the cost of reprepping those samples is NOT included in the quoted cost. For some experiments, repreps can be done by hand, but at a higher rate. Second, we are focused far more on reducing price than for standard library preparation, so some "routine services" - such as quality control or arraying of samples - are not built in. These services are available as add ons.
FRAGMENTED DNA
|
NEB UltraII is used for standard LM-PCR based approaches. We have miniaturized the reaction to a 1/5th volume with good performance on inputs from 10pg to 10ng (though we recommend a minimum of 100pg). The reaction begins with 10ul of sample to which reagents are added. As such, having clean initial samples in the exact correct volume is critical for the prep. |
INTACT GENOMES / TAGMENTATION
|
To minimize library prep costs for standard Illumina libraries, we have automated Illumina NexteraXT on our Tecan EVO150s and on our TTP Mosquito. Libraries will be built to include variable i5 and i7 indexes based on the grid so dual indexing is required during sequencing of these libraries. For more information about how Nextera works, please check out the standard Nextera library preparation section of standard DNA library prep. Libraries prepared on the Mosquito use a significantly lower volume for preparation. This allows for significantly reduced costs but also lowers the complexity. As such, is it most suitable for single cell and amplicon analysis. De novo work should not be done using the reduced volume MosquitoXT preps. We are also working on miniaturization of the Nextera FLEX system from Illumina. Initial results are promising. Once Illumina has larger index sets available, we will likely convert most NexteraXT preps to NexteraFLEX. |
16S / AMPLICON SEQUENCING
|
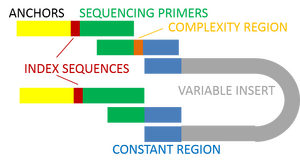
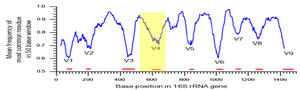
The BioMicro Center offers 16S and amplicon sequencing for metagenomics projects. Derived from the Alm Lab with support from a pilot grant from MIT CEHS, our protocol uses a two step amplification to first expand the 16S population and add defined 3' and 5' sequences which are then used to add Illumina anchors and sequences. This two step method allows easy multiplexing and the ability change the amplicon insert sequence at minimal cost. The same method used for 16S is also applicable to other amplicons as well with minor adaption. Because we use a nested PCR, only the internal sequences need to be modified. The adapter sequences - Green:Blue - are the key element of this method. On the 5' end, they contain a "YRYR" sequence that introduces the complexity required for Illumina sequencing. FORWARD: ACACGACGCTCTTCCGATCTYRYRXXXXXXXXXX (X = insert element) REVERSE: CGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATCTXXXXXXXXXXXX The most common source of failures for amplicon sequencing are samples that fail to amplify in the initial qPCR. The first step of the process is a qPCR using the forward and reverse primers to determine the number of cycles to be used in library generation. Libraries that fail to amplify in 20 cycles generally preform poorly enough to be unusable. Removal of PCR inhibitors is critical to success of this protocol. We do attempt to do this by dilution using a serial dilution in the initial qPCR test. The BioMicro Center generally recommends 250PE or 300PE MiSeq kits for sequencing 16S libraries.
|
 |
