Samantha M. Hurndon Week 3
From OpenWetWare
Jump to navigationJump to search
ACTIVITY ONE/PART TWO
METHODS
- I first started by going to pub med and searching for the article; Patterns of HIV-1 evolution in individuals with differing rates.
- From there I scrolled down to where I could the Secondary Source ID. Under this section there were plenty of HIV sequences, in which one was chosen (GenBank: AF016763.1)
- I Then wanted to down load several sequences in FASTA format. To do this I clicked on Related Sequences. Here you can see all the related sequences and click on several at one time. I selected 4.
RESULTS
- The accession number of the sequence I chose was AF016763.1
- The subject was from HIV-1 subject 1 visit 1 clone 1 from USA.
ACTIVITY ONE/PART THREE
- We used biology Workbench to analyze sequence data.
METHODS
- First I logged onto biology workbench, by creating an account.
- Once I was logged on there was five buttons that would take us to different tool sets. For this project, we are interested in nucleic sequence data.
- In the box with different tools I then selected; add new sequence. Then, pressed the “run” button.
- To add new sequences, I did so by uploading a file from my computer.
- After uploading my file, I pressed the Save button which then imported the data to biology work bench. All the sequences then appeared.
- Out of the sequences that appeared I then chose one sequence.
- Our next step after introducing ourselves to the biology workbench, was to highlight the ClustalW tool in the tool section.
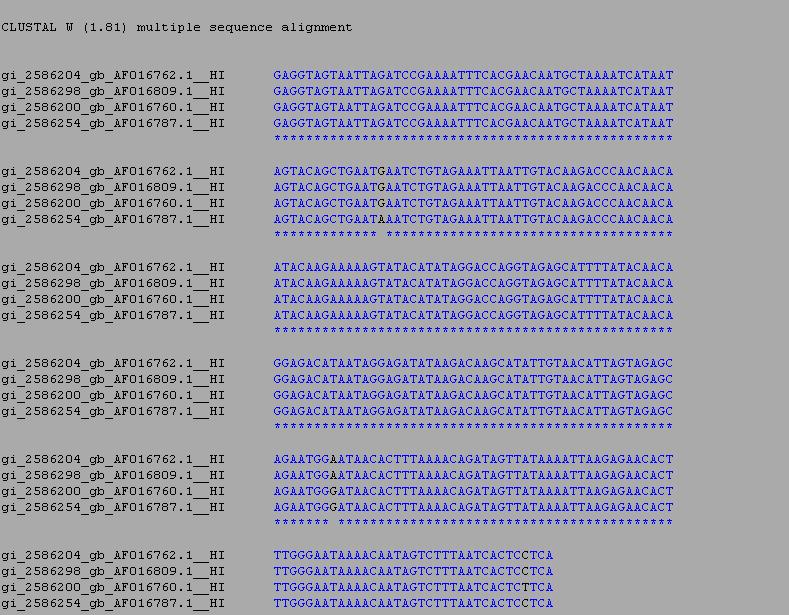
- From there we selected all of our sequences and ran a multiple sequence alignment using the ClustalW.
RESULTS
- Image of sequence alignment
ACTIVITY TWO/PART ONE
- For this activity we worked with HIV Sequence data that was collected from 15 individuals. The goal here is to analyze the isolated HIV and determine if they originated from a common source.
METHODS
- I first uploaded a file that contained sequencing onto the nucleic acid tool set of the biology workshop.
- Then, I generated a multiple sequence alignment and distance tree for 12 sequences (3 clones from each of the 4 subjects, which can be seen in the figure below)
RESULTS
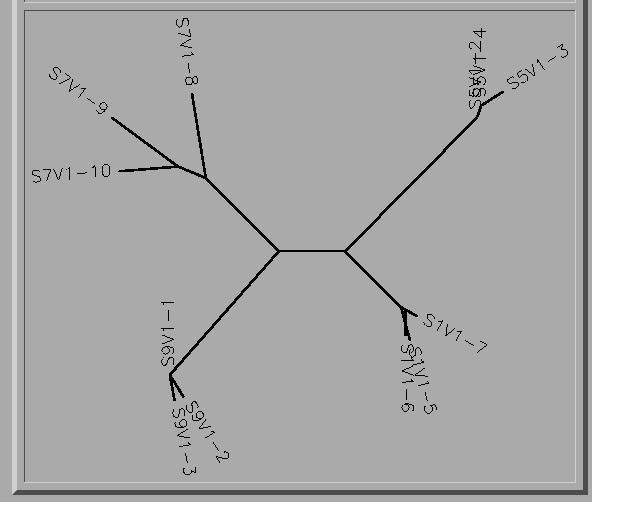
- All the clones from each subject do cluster together, some more than others.
- Subject Seven shows much more diversity than the other three subjects.
- Because the Central branch is so short in comparison to the branches that branch off, this is suggesting that we can not make any evolutionary assumptions between those of each subject. Some clones in each subject have much more in common than others. For example; S5V1-2 and S5V1-3 are basically on top of each other indicating they are very alike.
ACTIVITY TWO/PART TWO
- Here we looked at different ways to quantify sequence similarity and differences.
=METHODS
- First, Select all the clones from one subject (I used clone 8).
- Then calculate S by counting the number of disagreements in the nucleotides. (For clone 8 my S value was 6)
- Next, we were to calculate sigma using a online calculator tool given to us.
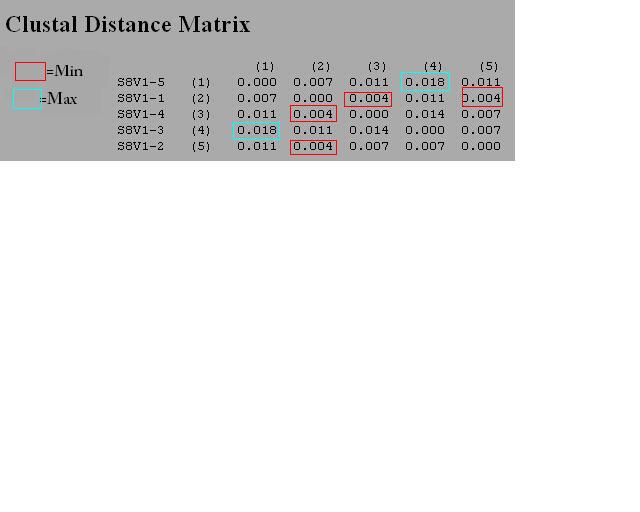
- Then, we used the Clustdist tool to generate a distance matrix for our alignment. Here we can find the min difference and max difference.
- Do this two more times and enter in a chart (See figure one under results for this information)
- The second part of this was to create a new alignment with all of the sequences from two subjects.
- We used the Clustdist once again to compare distances in nucleotides.
- This was done three times and entered in a table. (See figure two for data information, under results)
RESULTS
Interesting Links
Bioinformatics Individual Journal Entries
Week 6: DNA Glycosylase Exercise
Week9: Amino Acid Sequences/Presentation
Week 10: DNA Microarray Introduction
Samantha M. Hurndon Week 11: Week 11: DNA Microarray Journal Club
Samantha M. Hurndon Week 12: DNA Microarray project
Samanhta M. Hurndon Week 14: Final Project
Class Assignments