Samantha M. Hurndon Week 8
Group project
My partner for this research project is User: Nicolette S. Harmon. We will be examining the structures of the nonprogressor types and some of the moderate progressors from the markham study we used during the HIV Evolution project. The specific subjects we will be examining are 2,5,7,8,9,12,13,14 as they are subjects who were either classified as moderate progressors or nonprogressors. We will use the Star Biochem tool to analyze the protein structures.
Chapter 2
- Started by going to www.expasy.org/sprot/. Which is the Swiss=Prot database home page.
- Then typed in the search area for HIV gp120
- A list of relevant protein sequences appeared and I clicked on P04578 (ENV_HV1H2)
- I then scrolled down to look at the sequences, by clicking on FASTA it gave me the format seen below:
- >sp|P04578|ENV_HV1H2 Envelope glycoprotein gp160 OS=Human immunodeficiency virus type 1 group M subtype B (isolate HXB2) GN=env PE=1 SV=2
MRVKEKYQHLWRWGWRWGTMLLGMLMICSATEKLWVTVYYGVPVWKEATTTLFCASDAKA YDTEVHNVWATHACVPTDPNPQEVVLVNVTENFNMWKNDMVEQMHEDIISLWDQSLKPCV KLTPLCVSLKCTDLKNDTNTNSSSGRMIMEKGEIKNCSFNISTSIRGKVQKEYAFFYKLD IIPIDNDTTSYKLTSCNTSVITQACPKVSFEPIPIHYCAPAGFAILKCNNKTFNGTGPCT NVSTVQCTHGIRPVVSTQLLLNGSLAEEEVVIRSVNFTDNAKTIIVQLNTSVEINCTRPN NNTRKRIRIQRGPGRAFVTIGKIGNMRQAHCNISRAKWNNTLKQIASKLREQFGNNKTII FKQSSGGDPEIVTHSFNCGGEFFYCNSTQLFNSTWFNSTWSTEGSNNTEGSDTITLPCRI KQIINMWQKVGKAMYAPPISGQIRCSSNITGLLLTRDGGNSNNESEIFRPGGGDMRDNWR SELYKYKVVKIEPLGVAPTKAKRRVVQREKRAVGIGALFLGFLGAAGSTMGAASMTLTVQ ARQLLSGIVQQQNNLLRAIEAQQHLLQLTVWGIKQLQARILAVERYLKDQQLLGIWGCSG KLICTTAVPWNASWSNKSLEQIWNHTTWMEWDREINNYTSLIHSLIEESQNQQEKNEQEL LELDKWASLWNWFNITNWLWYIKLFIMIVGGLVGLRIVFAVLSIVNRVRQGYSPLSFQTH LPTPRGPDRPEGIEEEGGERDRDRSIRLVNGSLALIWDDLRSLCLFSYHRLRDLLLIVTR IVELLGRRGWEALKYWWNLLQYWSQELKNSAVSLLNATAIAVAEGTDRVIEVVQGACRAI RHIPRRIRQGLERILL
- I then went to the browser address www.expasy.org/sport/ and clicked on advanced search.
- from there I typed in HIV-1 V3 region. A list of results showed and I clicked on Q70446 (Q70446_9HIV1)
- The structure found here is bellow:
- >tr|Q70446|Q70446_9HIV1 Envelope glycoprotein, v3 region (Fragment) OS=Human immunodeficiency virus 1 GN=env PE=4 SV=1
CTRPNNNTRKSIRIGPGQTFYATGDIIGDIRQAHC
- I then did an advanced search for HIV gp120 and clicked on Q70442 (Q70442_9HIV1)
- Clicked on fasa again and got:
- >tr|Q70442|Q70442_9HIV1 Envelope glycoprotein, v3 region (Fragment) OS=Human immunodeficiency virus 1 GN=env PE=4 SV=1
- Clicked on fasa again and got:
CTRPNNNTRKSIRIGPGQTFYATGDIIGDIRQAHC
Chapter 4
- Started by going to www.expsy.ch/sprot/.
- From there I typed in HIV gp120 in the search area.
- I clicked on P03377
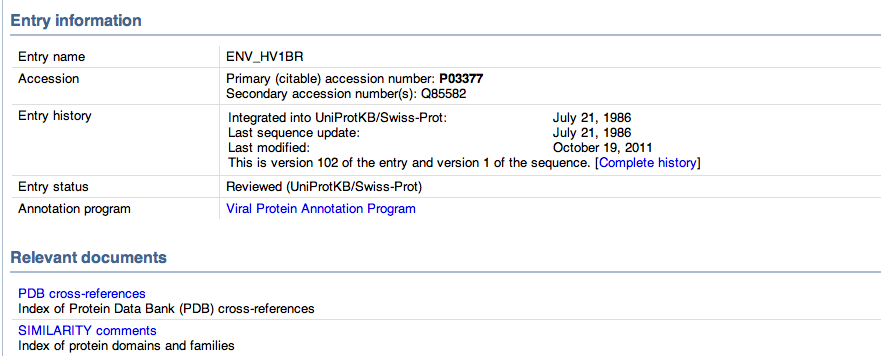
- At the bottom of the page the entry information is seen. Shown below
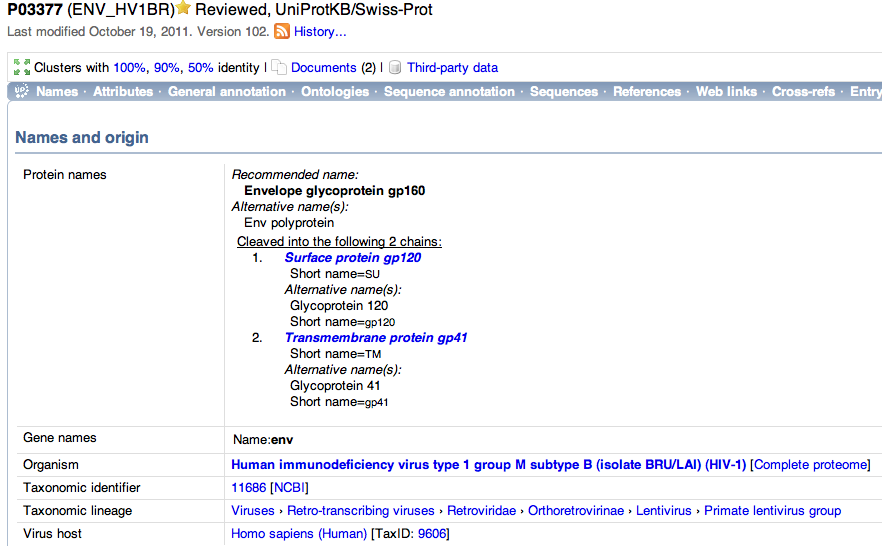
- I then went to the name and origin of the protein which was located at the top of the page and can be seen below
- I next went to the references located near the bottom of the page. This gave me several references , some seen below:
- The comments section was examined next and included information on function, subunit structure, subcellular location, domain, post-translational modification, miscellaneous and sequence similarities
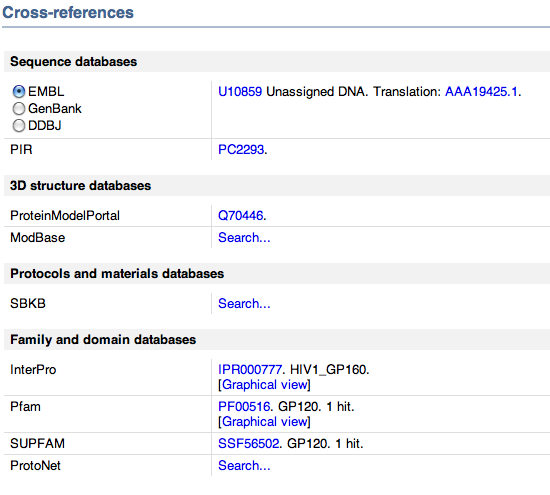
- Next, I went to cross references shown below. This area contains links to entries in other data bases, providing us with more information about our protein.
- Keywords: This gives relevant terms to the protein of inters along with the definition if you click on it
- The Features: This contains information on the protein that is mapped onto the sequence.
- The signal are numbers associated with out term
- The chain shows us the mature peptidic chain
- TOPO_DOM: is the topological domain
Chapter 5
- Here we went over ORFing our DNA sequences:
- In order to code for a protein the DNA sequence needs to have a translational Start codon (ATG) and not have any stop codons (TAA, TAG, TGA). Proteins have an average length of 350 residues.
- To get practice in ORFing I did the following:
- First, I went to www.ncbi.nlm. Nih.gov/gorf/gorf.html
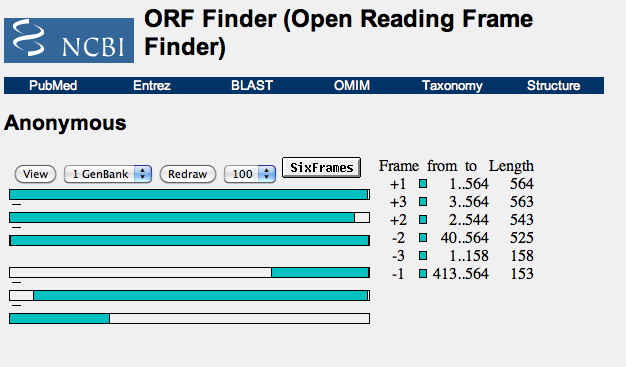
- I then input one of the sequences I found above input into the input box. What was found is shown below:
- Seen here is the DNA sequence which is displayed as six parallel horizontal bars and corresponds to one of six translational frames
- good for finding protein c oding regions for higher organisms if your sequence DNA is cDNA or mRNA.
- cDNA don’t include introns and have a simple microbe like ORF structures.
Chapter 6: Working with a Single Protein Sequence
- important question to ask yourself: what does your protein look like and when is it active
- Does it need to be modified after translated?
- Does it contain coiled-coil elements?
- Is it a transmembrane protein?
- to help you in guessing the function of a protein you can perform similarity searches. (finding a similar protein in a database )
- important question to ask yourself: what does your protein look like and when is it active
- Predicting the main physico-chemical properties of a protein:
- First I pointed my browser to www.expasy.org/tools/#primary
- I then clicked on the protparam and entered my sequence in the search boxes
- What was found can be located on this web page http://web.expasy.org/cgi-bin/protparam/protparam
- Digesting a protein in a computer
- Protease digestions is where you use an enzyme to cut your protein
- Can be found at www.expasy.org/tools/#proteome
- Doing primary Structure Analysis
- amino acid sequence of the protein done to find segments of the protein that exhibits special conditions.
- Running Protscale
- I first went to www.expasy.org/cgi-bin/protscale.pl.
- Then entered my sequence number
- After the sequence number was entered I then scrolled clicked the radio button and chose 19 from the pull down menu.
- My findings from this are seen here: http://web.expasy.org/cgi-bin/protscale/protscale.pl?1
- Running TMHMM
- This predicts transmembrane regions
- I went to www.cbs.dtu.dk/services/TMHMM-2.0
- Results can be seen here http://www.cbs.dtu.dk/cgi-bin/nph-webface?jobid=TMHMM2,4EA387C5011395A2&opt=none
- This predicts transmembrane regions
- PROSITE patterns here you can compare proteins with a list of proteins that are in the database
- you can do this by goin to www.expasy.org/tools/scanprosite/. Pasting your sequence, unchecking the exclude motifs box and press start scan
- Results of this can be seen here http://prosite.expasy.org/cgi-bin/prosite/ScanView.cgi?scanfile=341454828621.scan.gz