IGEM:IMPERIAL/2006/project/Oscillator/project browser/Predator Construct/Modelling
From OpenWetWare
Jump to navigationJump to search
| Super Parts | Molecular Prey-Predator Oscillator | |
|---|---|---|
| Actual Part | 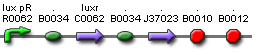
| |
| Sub Parts | Test Sensing Predator Construct | Test Killing Predator Construct |
Model assumptions and relevance
- General assumptions on gene expression modelling:
- Quasi-steady state hypothesis on mRNA expression.
- Gene activation can be approximated by Hill equations.
- Assumptions linked to the quorum sensing:
- As a first approximation, we assume that luxR and AHL molecules form a heterodimer (even if it has been found that the complex formed is more complicated).
- The degradation rate of luxR and aiiA is mainly due to the growth dilution.
<html> <script type="text/javascript" language="javascript"> var sc_project=1999441; var sc_invisible=1; var sc_partition=18; var sc_security="18996820"; </script>
<script type="text/javascript" language="javascript" src="http://www.statcounter.com/counter/frames.js"></script><noscript><a href="http://www.statcounter.com/" target="_blank"><img src="http://c19.statcounter.com/counter.php?sc_project=1999441&java=0&security=18996820&invisible=1" alt="website statistics" border="0"></a> </noscript> </html>
Model description of the growth of the predator
- mathematical description of the predator growth and death:
- [math]\displaystyle{ \frac{d[luxR]}{dt} = \frac{c * [AHL] * [luxR]}{(c0 + [AHL] * [luxR])} - gd * [luxR] }[/math]
- [math]\displaystyle{ \frac{d[aiiA]}{dt} = \frac{c * [AHL] * [luxR]}{(c0 + [AHL] * [luxR])} - gd * [aiiA] }[/math]
- insert a graphical representation if possible (e.g. CellDesigner display)
- link to SBML file or matlab.
Model variables and parameters for the growth of the predator
(list all the variables and parameters of the model in a table, specifying if their values are known, unknown, to be measured.)
| Variables | ||||
| Name | Description | Initial Value | Confidence | Reference |
|---|---|---|---|---|
| AHL | homoserine lactone acting as the prey-molecule | 0 | depends how good is the control of the prey positive feedback | links |
| luxR | molecule acting as the sensing module for the predator | 0 | to be measured as we might have to deal with some leakage of the promoter | links |
| aiiA | molecule acting as the killing module of the prey for the predator | 0 | to be measured as we might have to deal with some leakage of the promoter | links |
| Parameters | ||||
| Name | Description | Value | Confidence | Reference |
|---|---|---|---|---|
| c | maximum synthesis rate of the pLux promoter | to be characterized | to be measured | links |
| c0 | dissociation constant according to Hill eq | to be characterized | to be measured | links |
| gd | growth dilution | to be characterized | to be measured | links |
Model description of the killing of the prey molecule by the predator
- mathematical description of the killing of the prey:
- [math]\displaystyle{ \frac{d[AHL]}{dt} = \frac{b * [aiiA] * [AHL]}{(b0 + [AHL])} - e * [AHL] }[/math]
Model variables and parameters for the killing of the prey molecule by the predator
| Variables | ||||
| Name | Description | Initial Value | Confidence | Reference |
|---|---|---|---|---|
| AHL | homoserine lactone acting as the prey-molecule | 0 | depends how good is the control of the prey positive feedback | links |
| aiiA | molecule acting as the killing module of the prey for the predator | 0 | to be measured as we might have to deal with some leakage of the promoter | links |
| Parameters | ||||
| Name | Description | Value | Confidence | Reference |
|---|---|---|---|---|
| b | Maximum degradation rate catalyzed by aiiA | ... | to be measured | links |
| b0 | Michaelis-Menten constant of enzyme reaction | ... | to be measured | links |
| e | AHL wash-out | variable | to be measured/can be varied by chemostat | links |
Dynamical and sensitivity analysis
<showhide>
- Template__HIDER__
<hide>
- analyze model in order to show how the part could fulfill its specifications
- insert graph and charts
</hide></showhide> <showhide>
- We have decoupled the predator into sensing and killing parts
- For detail modelling, please refer to the modelling section in subparts.
Characterization
- To characterize the part, we have to measure the production rate and aiiA enzyme activity seperately
- For details characterization, please refer to subparts