Cdominguez Week 13
From OpenWetWare
Jump to navigationJump to search
Methods/Results
- Converted the spike protein DNA sequence into a protein sequence using either the ExPASY Translate tool.
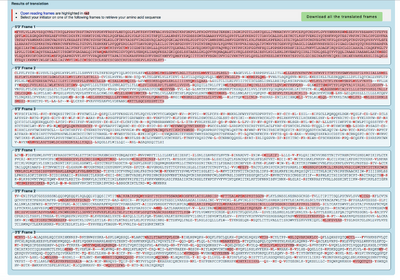
- How do you know which of the six frames is the correct reading frame (without looking up the answer)? Of the six frames, the correct reading frame is the one with the most part highlighted red so the first frame.
- Checked answer with the NCBI protein record. The answer is Frame 1
- Found out what is already known about the spike protein in the UniProt Knowledgebase (UniProt KB). UniProt KB has two parts to it, Swis-Prot, which contains entries for proteins that have been manually reviewed, and TrEMBL (which stands for "Translated EMBL"), which are automated translations of all DNA sequences in the EMBL/GenBank/DDBJ databases. SARS-CoV-2 is so new that it has not yet been added to the UniProt database; it is scheduled to be added with the April 22 release.
- If you search on the keywords "SARS-CoV" (which refers to the first SARS virus), in the main UniProt search field, how many results do you get? 818
- Used the entry with accession number "P59594" which corresponds to the reference entry for the SARS-CoV spike protein.
- What types of information are provided about this protein in this database entry? There are two spike proteins, S1 and S2. S1 attaches virus to cell membrane by host receptor. It states the ACE2 receptor and CLEC4M/DC-SIGNR receptors are the receptors in which it attaches. S2 mediates fusion as a class I viral fusion protein. It speaks to the 3 conformational states that occur during fusion. Post-translational modification includes the spike glycoprotein digested with host endosomes. They also have a secondary and tertiary structure model of the protein.
- We are going to use the PredictProtein server to analyze the SARS-CoV-2 spike protein.
- Pasted the SARS-CoV or SARS-CoV-2 spike protein amino acid sequence that was discussed in Wan et al. into the input field and submit
- Paste a screenshot of the results into your wiki. Note that you can zoom in on different parts of the protein by using the slider at the top. Explore the types of information provided (in the menu options at the left). How does this information relate to what is stored in the UniProt database for the SARS-CoV spike protein? In the paper, they specify that a histidine in place of lysine at 353 would mean that ACE2 is probably not used as the receptor like in the mouse and rate. I checked this point mutation to see what would happen. The software would not allow me to do this. It would load and freeze so that it reloaded the page and I was not able to see individual point mutations. I did not see any information about the S1 and S2 receptors.
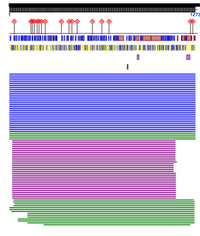
Predict Protein Server Results
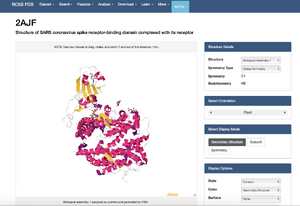
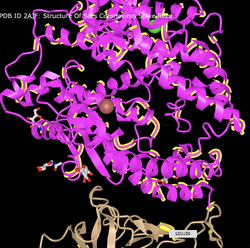
- View the structure of the SARS-CoV or SARS-CoV-2 spike protein from your assigned journal club article at the Protein Data Bank.
- Wan et al. (2020): 2AJF
- Clicked on the "Structure" underneath the image of the structure on the upper left side of the page.
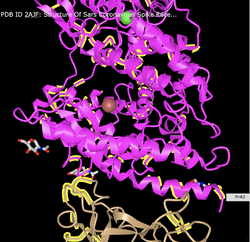
- Alternately, NCBI has a web-based structure viewer called iCn3D. To use it:
- Went to the NCBI Structure home page and pasted 2AJF for my structure (above) into the search field and clicked "search".
- On the results page, clicked on the button for the "full-featured 3D viewer" found on the bottom right corner of the structure image.
- This opened up a window with a similar image viewer to the one found on PDB. The interface is a little clunkier and the images are less elegant, but the iCn3D viewer has the advantange of showing the structure in a way that makes it easier to identify the N- and C-terminus (there are arrows for the beta strands and the helices are also pointed). It also allows you to see the sequence at the same time and find and highlight particular sequences in the structure.
- When answering the following questions, provide a screenshot pasted into your wiki:
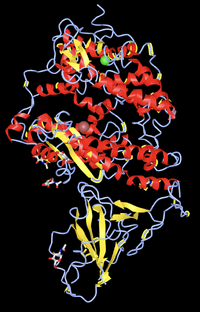
- Create a view of the protein from your paper recreates one of the figures from your paper. You may not be able to get this to be exactly the same in terms of colors or backbone style, but you should try to rotate the to be the same view as the article.
- Find the N-terminus and C-terminus of each polypeptide tertiary structure.
- Locate all the secondary structure elements. Do these match the predictions made by the PredictProtein server? I believe it matches the predictions made by the PredictProtein server. It stated that it was about 50% loop, 25% strand, and 25% helix. Looking at the structure, I would say this is true.'
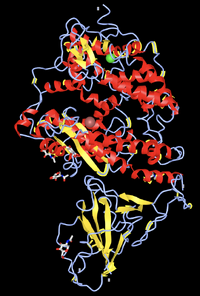
- Locate particular amino acids that were discussed in your paper, show a screenshot that highlights them. Phenylalanine 486 was said to form an unfavorable interaction with Threonine 82 due to a polar side chain. I was able to location those two amino acids relatively close to each other on what looks like the same part of the protein with different numbers (Threonine 2). This seems to locate that amino acid.I also looked for Leu455 and Glutamine493 since they lose a favorable interaction. I was also able to locate these on the protein with different amino acid numbers (Leucine 156).
Research Project Questions
With your new final project partners, answer the following:
- What question will you answer about sequence-->structure-->function relationships in a SARS-CoV-2 protein?
- Are there differences between the sequence of the RBDs of ACE-2 of SARS-CoV and SARS-CoV2 that potentially cause difference in binding strength?
- What sequences will you use?
- The sequences of ACE2 of SARS-CoV as well as SARS-CoV2.
- What protein tools will you use for analysis and answering your question?
- PredictProtein Server, Protein Data Bank, and NCBI Structure
Data and Files
Scientific Conclusion
The Wan et al. paper was mostly consistent with the data sources that were used. It was difficult to be able to translate from the paper to the structure databases mostly due to the difference in numbering of the amino acids. From lining up the images, the interactions of amino acids that were mentioned did also show in the structure databases further validating their paper and hypothesis. Overall, this exploratory assignment was difficult at times to translate from the paper into evaluation; however, the paper was successful at predicting the interaction of the receptor from a qualitative perspective.
Acknowledgments
- I worked with User:Sahil Patel and User:Adinulos where we met via Zoom to work on our research question.
- Except for what is noted above, this individual journal entry was completed by me and not copied from another source. Cdominguez (talk) 18:04, 21 April 2020 (PDT)
References
- Expasy. (2020). ExPASY Translate Tool. Retrieved April 21, 2020, from http://web.expasy.org/translate
- NCBI.(2020). NCBI Open Reading Frame Finder. Retrieved April 21, 2020, from https://www.ncbi.nlm.nih.gov/orffinder/
- NCBI. (2020). NCBI Structure Database. Retrieved April 21, 2020, from https://www.ncbi.nlm.nih.gov/Structure/index.shtml.
- OpenWetWare. (2020). BIOL368/S20:Week 13. Retrieved April 21, 2020, from https://openwetware.org/wiki/BIOL368/S20:Week_13.
- Predict Protein Server (2020). Predict Protein Server. Retrieved April 21, 2020, from https://ppopen.informatik.tu-muenchen.de/
- Protein Data Bank. (2020). Protein Data Bank. Retrieved April 21, 2020, from https://www.rcsb.org
- Uniprot. (2020). Uniprot. Retrieved April 21, 2020, from http://www.uniprot.org
- Wan, Y., Shang, J., Graham, R., Baric, R. S., & Li, F. (2020). Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. Journal of virology, 94(7). DOI: 10.1128/JVI.00127-20.