Adinulos Week 13
Template
User Page
Assignments
- BIOL368/S20:Week 1
- BIOL368/S20:Week 2
- BIOL368/S20:Week 3
- BIOL368/S20:Week 4
- BIOL368/S20:Week 5
- BIOL368/S20:Week 6
- BIOL368/S20:Week 8
- BIOL368/S20:Week 9
- BIOL368/S20:Week 10
- BIOL368/S20:Week 11
- BIOL368/S20:Week 13
- BIOL368/S20:Week 14
Journal Pages
- adinulos Week 2
- adinulos Week 3
- adinulos Week 4
- adinulos Week 5
- adinulos Week 6
- adinulos Week 8
- adinulos Week 10
- adinulos Week 11
- adinulos Week 13
- adinulos Week 14
Class Journal Pages
- BIOL368/S20:Class Journal Week 1
- BIOL368/S20:Class Journal Week 2
- BIOL368/S20:Class Journal Week 3
- BIOL368/S20:Class Journal Week 4
- BIOL368/S20:Class Journal Week 5
- BIOL368/S20:Class Journal Week 6
- BIOL368/S20:Class Journal Week 10
- BIOL368/S20:Class Journal Week 11
- BIOL368/S20:Class Journal Week 13
- BIOL368/S20:Class Journal Week 14
Purpose
To critically analyze the structure of the spike protein for SARS-COV-2 using multiple analysis tools.
Methods and Results
Exploring spike protein structure
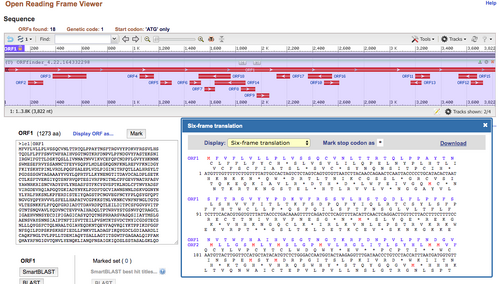
Convert the spike protein DNA sequence into a protein sequence using the NCBI Open Reading Frame Finder
- You know that the first reading frame (ORF1) is the correct reading frame because it shows the full sequence of the protein, the other reading frames are fragmented.
- This reading frame is correct because the sequence in ORF1 matches the sequence on the NCBI protein record for the spike protein
Figure 1. Reading frame results from NCBI Open Reading Frame Finder
Use the UniProt Knowledgebase (UniProt KB) website to find out what kind of information on the protein is stored in the database
- 833 results are found when you search "SARS-CoV".
- Information provided in the protein with the ascension number "P59594" includes information about the function of the protein, the names and taxonomy of the protein, the subcellular location, pathology and biotech, PTM/Processing, the interaction of the protein, the structure of the protein, the sequence, a list of similar proteins, cross-references and entry information.
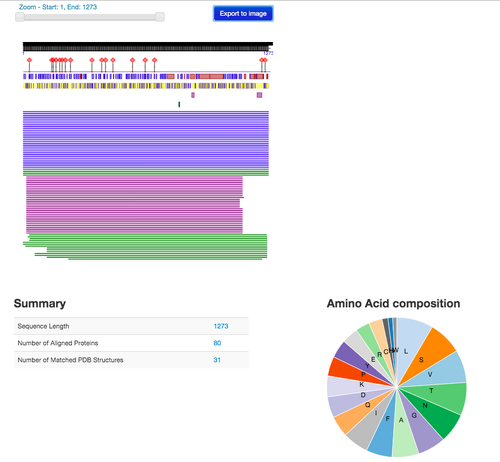
Use the PredictProtein server to analyze the SARS-CoV-2 spike protein using the amino acid sequence from the Wrapp paper. Paste the sequence into the input field
- The information provided on the PredictProtein website includes the structure annotation, function annotation, and additional services. This information is similar to what is included on the UniProt page, however, it seems that UniProt may include this information with more detail.
Figure 2. PredictProtein results using Wrapp amino acid sequence.
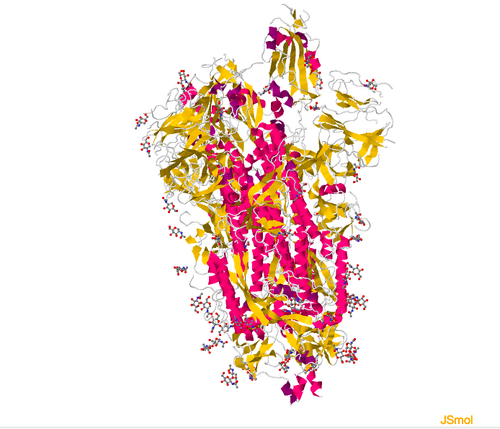
View the structure of the SARS-CoV-2 spike protein from the Wrapp paper, 6VSB, click "Structure" underneath the image of the structure on the upper left side of the page. This will open a winder where you will be able to interact with the structure of the image. Go to "Select a different viewer, and choose Jsmol,
- Recreate the view of the protein included in the Wrapp et. al paper.
- Use the NCBI iCn3D viewer, to get an in-depth view of the sequences.
- The N-terminii are located and the C terminii were located for each polypeptide structure using the green arrows to indicate the start and end of the helices and sheets.
- These locations may match up with the ProteinPredictor results; however, it is hard to determine this by the image. Using the slider bar to zoom into the ProteinPredictor shows similarities between the locations with the NCBI produced structure.
Figure 3. Structure of the Wrapp et. al spike protein
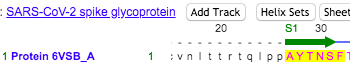
- Locations of the NTD were found in the sequence on the NCBI structure that was highlighted in the Wrapp paper. See Figures 4 and 5 below for similar amino acid sequences.
Figure 4. NTD Sequence obtained from the NCBI 3D imaging.
Figure 5. Sequence for NTD derived from the Wrapp et. al paper.
Beginning research project
- My partners and I were curious about the specific sequence of the RBD of the spike protein. Are there any differences between the sequence of the RBD's for ACE-2 of the SARS-CoV and SARS-CoV-2 that potentially cause SARS-CoV-2 to bind more tightly than SARS-CoV?
- We would use the spike protein sequences for SARS-CoV and SARS-CoV-2 and highlight differences specifically in the RBD region of the protein. To analyze we would use a sequence aligner, open reading frame finder, and the protein predictor.
Data and Files
Figure 1 Figure 2 Figure 3 Figure 4 Figure 5
Conclusion
The structure of the spike glycoprotein for the SARS-CoV virus was analyzed. Various tools were used to determine areas of specific amino acid sequences that were highlighted in the Wrapp. et al paper.
Acknowledgments
- I communicated with my partners, Christina and Sahil to develop our project idea.
- Protocol was taken from the Week 13 Wiki.
- Except for what is noted above, this individual journal entry was completed by me and not copied from another source.
Adinulos (talk) 17:56, 22 April 2020 (PDT)
References
NCBI. (2020). ORFfinder. Retrieved 22 April 2020, from https://www.ncbi.nlm.nih.gov/orffinder/
NCBI. (2020). Structure. Retrieved April 22 2020, from https://www.ncbi.nlm.nih.gov/Structure/index.shtml
OpenWetWare. (2020). BIOL368/S20:Week 13. Retrieved April 22, 2020, from https://openwetware.org/wiki/BIOL368/S20:Week_13
PredictProtein. (2020). open.predictprotein.org. Retrieved April 22, 2020, from https://open.predictprotein.org
RCSB. (2020). 6VSB. Retrieved April 22 2020, from https://www.rcsb.org/pdb/explore/jmol.do?structureId=6vsb&bionumber=1&jmolMode=HTML5
UniProt. (2020). Uniprot.org. Retrieved 22 April 2020, from https://www.uniprot.org/
Wrapp, D., Wang, N., Corbett, K. S., Goldsmith, J. A., Hsieh, C. L., Abiona, O., ... & McLellan, J. S. (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science, 367(6483), 1260-1263.