User:Anthony Salvagno/Notebook/Research/2010/02/16/More Poster Design
Quick Navigation: Pages by Category | List of All Pages | Protocols
Tentative Plan
<html> <iframe width="800" height="600" frameborder="1" scrolling="no" src="http://www.mindmeister.com/maps/public_map_shell/42141568/Ant's-2010-Biophys-Poster?width=800&height=600&zoom=1&live_update=1" style="overflow:hidden"></iframe> </html>
Design
I'm thinking of doing a tree like structure to demonstrate the mindmap, with extraneous stuff (like acknowledgments, future work, etc) at the bottom.
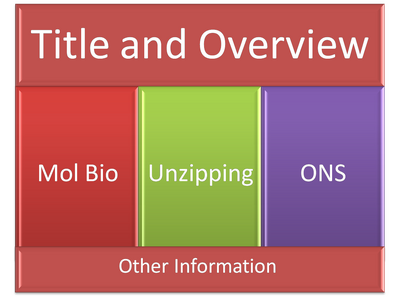
This is my idea quickly in powerpoint. But not as colored. I was thinking something like a subway map design. We'll see...
Content
I will gether notebook information for this poster and put it all here. I will make my poster in Illustrator and will edit and compile it later.
Submitted to BPS
Title:APPLICATION OF SHOTGUN DNA MAPPING TO YEAST GENOMIC DNA SHOTGUN CLONES
Abstract:Shotgun DNA mapping (SDM) is the ability to identify the genomic location of a random DNA fragment based on its naked DNA unzipping forces compared with simulated unzipping forces of a published genome. We have previously demonstrated proof of principle for shotgun DNA mapping by using plasmid pBR322 unzipping data amongst yeast genome background[cite preprint]. Currently we are validating the technique using unzipping data from yeast genomic DNA. Genomic DNA from yeast (S. cerevisiae) has been digested with restriction endonucleases to produce a library of random fragments, which we used to create a limited library of shotgun clones. Single-molecule unzipping constructs derived from these clones will be unzipping with optical tweezers (OT). In parallel, we have created a library of simulated possible unzipping force profiles, based on the known yeast genome sequence. The OT data and the library will be used in our existing SDM algorithms to identify each shotgun clone, and success rate will be determined via DNA sequencing of the clones. A major application of SDM we are working towards is mapping of nucleosomes and RNA Polymerase II molecules on native chromatin. We will report our progress towards this goal and also discuss other applications of SDM, including splice variant and telomere analysis.
That title is pretty gay. Can I change my title slightly? How bout Shotgun DNA mapping via yeast genomic DNA?
Project Overview
I would like to keep this short and sweet: It is possible to distinguish genomic information based on unzipping DNA using optical tweezers. Because of this, we get an unzipping signature primarily due to the base-pairing of DNA. We can use this information to match an unzipped sequence to a library of sequences obtained through simulations. We call this technique DNA Mapping and have found that we can use it to better understand protein-DNA interactions and the interaction locations for applications in chromatin mapping and structural DNA mapping.
I think this is a decent start. Maybe I should have some sort of ending line that ends in "..."
Molecular Biology
Here I would like to discuss the process of getting Shotgun clones. Steps include:
- digested genomic DNA
- we used XhoI and EcoRI
- cloning random fragments
- cloned in pBS KS-
- extracted 5 different clones and created "library" of random clones
- Show this picture (the second one)
- ligating clones to unzipping construct
- digest with SapI
- create unzipping construct
- PCR a 1.1kb fragment
- anneal short duplex oligo
- 3-piece ligation
- Definitely show this image for proof of ligation success.
- maybe a side-by-side comparison of this image and this image. The later will have to be heavily Photoshopped.
- also tried ligating cap to hinder concatamers
Unzipping Experiments
- Optical Tweezers
- image of the current setup with caption that describes setup
- maybe also optical path image
- image of the current setup with caption that describes setup
- Tethering
- components needed
- how it works - my image of unzipping or remake Diego's old image
- maybe flip book of tethered particle motion
- my technique or protocol and what I look for in a successful run
- Software
- I think just an image of comparison should do the trick here
Open Science
I won't say all of the following, maybe images will suffice for some parts, but I may easily talk about all of it during my presentation
- Open Notebook
- OpenWetWare - completely editable wiki allows sharing data in multiple ways
- post protocols
- post methods
- post failures and success
- easy access for other lab members and anyone (Google searchable)
- "publish" data
- Google Docs
- can share with the world, one person, no one
- useful for templates and reproducible experiments
- accessible from anywhere
- OpenWetWare - completely editable wiki allows sharing data in multiple ways
- Open Data
- via notebooks
- via kochlab.org
- Open Proposals and Papers
- Open Proposal
- I don't know what to say here
- Open Papers (coming soon)
- Completely commentable
- Open Proposal
- Using Technology
- improves collaborations: FriendFeed/Facebook, Blogger
- improves data exposure: Youtube, Jove, Slideshare
- improves teaching: slideshare, youtube, blogger, etc