HughesLab:JTK Cycle: Difference between revisions
No edit summary |
No edit summary |
||
| Line 7: | Line 7: | ||
<p style="width:750px;"> | <p style="width:750px;"> | ||
JTK_Cycle is a non-parametric algorithm developed in collaboration with [http://scholar.google.com/citations?user=1hw5bX4AAAAJ&hl=en Professor Karl Kornacker], Ohio State University, and [http://bioinf.itmat.upenn.edu/hogeneschlab/index.php Professor John Hogenesch], University of Pennsylvania School of Medicine. Its purpose is to identify rhythmic components in large, genome-scale | JTK_Cycle is a non-parametric algorithm developed in collaboration with [http://scholar.google.com/citations?user=1hw5bX4AAAAJ&hl=en Professor Karl Kornacker], Ohio State University, and [http://bioinf.itmat.upenn.edu/hogeneschlab/index.php Professor John Hogenesch], University of Pennsylvania School of Medicine. Its purpose is to identify rhythmic components in large, genome-scale | ||
data sets, and estimate their period length, phase, and amplitude.</p> | |||
<p style="width:750px;"> | <p style="width:750px;"> | ||
JTK_Cycle runs in [http://en.wikipedia.org/wiki/R_%28programming_language%29 R], a statistical language that can be downloaded [http://www.r-project.org/ here]. | JTK_Cycle runs in [http://en.wikipedia.org/wiki/R_%28programming_language%29 R], a statistical language that can be downloaded [http://www.r-project.org/ here]. The scripts necessary to run JTK_Cycle are available as a .zip: [[Image:JTK_Cycle.zip]]. This compressed file also contains a User's Guide and several example data sets. | ||
</p> | </p> | ||
Revision as of 18:12, 8 January 2013
Home Lab Members Research JTK_Cycle MuscleDB Publications Protocols Academic Analytics Open Positions Contact
JTK_Cycle
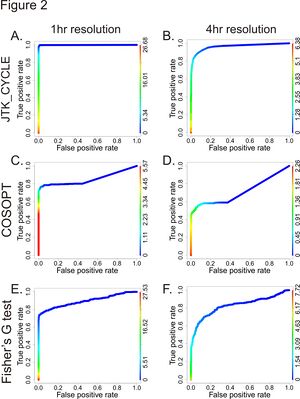
ROC curves showing that JTK_Cycle out-performs COSOPT and Fisher's G-test.
JTK_Cycle is a non-parametric algorithm developed in collaboration with Professor Karl Kornacker, Ohio State University, and Professor John Hogenesch, University of Pennsylvania School of Medicine. Its purpose is to identify rhythmic components in large, genome-scale data sets, and estimate their period length, phase, and amplitude.
JTK_Cycle runs in R, a statistical language that can be downloaded here. The scripts necessary to run JTK_Cycle are available as a .zip: File:JTK Cycle.zip. This compressed file also contains a User's Guide and several example data sets.

Hughes Lab
Department of Biology
University of Missouri, St. Louis
(Starting in August, 2013)
