HughesLab:Research
Home Lab Members Research JTK_Cycle MuscleDB Publications Protocols Academic Analytics Open Positions Contact
Transcriptional profiling of skeletal muscle
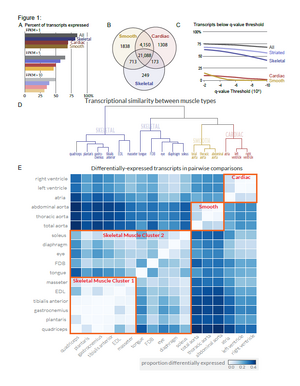
Transcriptional profiling reveals extraordinary diversity among skeletal muscle tissues. Terry et al. (2018)
Skeletal muscle comprises a family of diverse tissues with highly specialized morphology, function, and metabolism. Many acquired diseases, including HIV, COPD, cancer cachexia, critical illness myopathy, and sepsis, affect specific muscles while sparing others. Even monogenic muscular dystrophies tend to selectively affect certain muscle groups, despite their causative genetic mutations being present in all tissues. These observations suggest that factors intrinsic to muscle tissues influence their susceptibility to various disease mechanisms. Nevertheless, most studies have not addressed transcriptional diversity among skeletal muscles. Here we use unbiased RNA sequencing (RNA-seq) to profile global mRNA expression in a wide array of skeletal, smooth, and cardiac muscle tissues from mice and rats. Our data set, MuscleDB, reveals extensive transcriptional diversity, with greater than 50% of transcripts differentially expressed among skeletal muscle tissues. This diversity is only partly explained by fiber type composition and developmental history, suggesting that specialized transcriptional profiles establish the functional identity of muscle tissues. We find conservation in the transcriptional profiles across species as well as between males and females, indicating that these data may be useful in predicting gene expression in related species, such as humans. Notably, thousands of differentially expressed genes in skeletal muscle are associated with human disease, and hundreds of these genes encode targets of drugs on the market today. Related to this observation, we suggest a mechanistic explanation for how myotonic dystrophy induces weakness in the extensor digitorum longus (EDL) while sparing nearby muscles. These data may therefore provide the means by which muscle-specific sensitivity to disease may be unraveled. In addition, we detect hundreds of putative myokines that may underlie the endocrine functions of skeletal muscle. We anticipate that in conjunction with transcriptional modeling, this resource will catalyze more sophisticated tissue engineering of skeletal muscle to improve the efficacy of regenerative medicine.
ExpressionDB
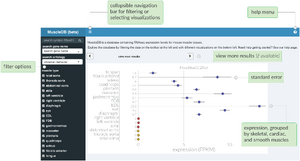
RNA-sequencing (RNA-seq) and microarrays are methods for measuring gene expression across the entire transcriptome. Recent advances have made these techniques practical and affordable for essentially any laboratory with experience in molecular biology. A variety of computational methods have been developed to decrease the amount of bioinformatics expertise necessary to analyze these data. Nevertheless, many barriers persist which discourage new labs from using functional genomics approaches. Since high-quality gene expression studies have enduring value as resources to the entire research community, it is of particular importance that small labs have the capacity to share their analyzed datasets with the research community. Here we introduce ExpressionDB, an open source platform for visualizing RNA-seq and microarray data accommodating virtually any number of different samples. ExpressionDB is based on Shiny, a customizable web application which allows data sharing locally and online with customizable code written in R. ExpressionDB allows intuitive searches based on gene symbols, descriptions, or gene ontology terms, and it includes tools for dynamically filtering results based on expression level, fold change, and false-discovery rates. Built-in visualization tools include heatmaps, volcano plots, and principal component analysis, ensuring streamlined and consistent visualization to all users. All of the scripts for building an ExpressionDB with user-supplied data are freely available on GitHub, and the Creative Commons license allows fully open customization by end-users. We estimate that a demo database can be created in under one hour with minimal programming experience, and that a new database with user-supplied expression data can be completed and online in less than one day.
Understanding the mechanism and physiological significance of downstream CCG regulation
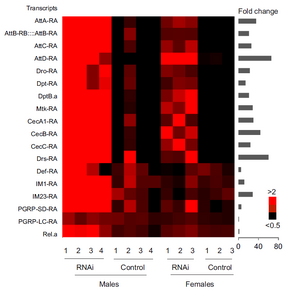
RNA-seq data showed an activation of immune responsive genes in Achl RNAi flies.
One of the most important questions to be addressed in the field is to identify CCG (Clock Controlled Gene)s that mediate clock output of physiological processes and to understand how they mediate clock output of physiological processes.
Here we found Achilles (Achl), a previously uncharacterized CCG, is rhythmically expressed in the fly brain, and Achl is likely to mediate the rhythmic immune response in flies. We found that the expression of Achl in neurons, represses the expression of immune responsive genes in the fat body (Figure 3) and consequently the resistance of flies against bacterial infection. Notably, we noticed a decreased overall lifespan and starvation resistance in Achl knockdown flies, suggesting a a behavioral or metabolic cost of constitutively activating immune pathways and furthermore underscores the significance of maintaining rhythmic homeostasis.
Ongoing work is focused on identifying the significance of Achl in the rhythmic regulation of immune system and deciphering the signaling mechanism downstream of Achl that conveys information from neurons to the fat body.
RNA-seq parameters in circadian rhythms
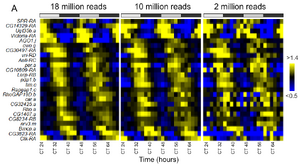
Many cycling transcripts are detectable with fewer reads per sample.
The development of next-generation sequencing, especially RNA-seq, has significantly accelerated circadian studies in profiling tissue-specific CCGs (clock controlled genes) as well as elucidating the mechanism of circadian output pathways. However, there are technical challenges await when conducting these experiments.
To address these challenges, we performed computational simulations and figured out the significance of some major challenges such as the depth of sequencing coverage, read-depth (See an example in Figure 4), and choice of alignment algorithms in using RNA-seq analysis in circadian rhythms using mice and Drosophila, two most commonly used model organisms.
Discovery of circadian harmonics
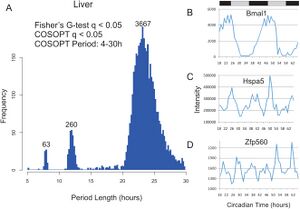
Transcriptional profiling of the mouse liver identified rhythmic transcripts with period lengths of ~8, ~12, and ~24 hours.
We profiled global gene expression over two full days using Affymetrix microarrays. We identified rhythmic transcripts in the mouse liver and pituitary, as well as fibroblasts (NIH3T3) and osteosarcoma cells (U2OS). These data have been made freely available on CircaDB as a resource to the field. To our surprise, we found several hundred genes cycling with period lengths much shorter than 24 hours (Figure 5). These ultradian rhythms had period lengths of ~8 and ~12 hours -- i.e., the second and third harmonics of 24 hour oscillations.
Subsequently, we have shown that these rhythms are found in tissues throughout the body. Moreover, they are found in fruit flies as well, suggesting that circadian harmonics are a common feature of animal transcriptional rhythms. At a mechanistic level, 12 hour rhythms require both a central and peripheral circadian oscillator, indicating that these rhythms are ultimately downstream of the conventional circadian clock. Typically, they are involved in cellular responses to stress, suggesting that ultradian transcriptional rhythms respond to twice daily stresses.
System-driven circadian oscillations
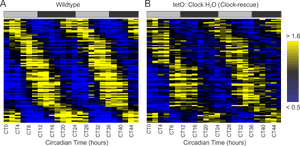
~100 genes oscillate in the mouse liver, despite ablation of the local, peripheral clock.
In collaboration with the Takahashi laboratory, we profiled transcriptional rhythms in mice with and without a defective circadian clock in the liver. Although most rhythmic genes are lost due to the ablation of the local circadian oscillator, nearly 100 genes continue to cycle with appropriate period lengths and phases (Figure 6). Consequently, these persistent rhythms may form the molecular basis by which the central clock drives peripheral oscillations. Strikingly, many core clock genes continue to oscillate in the absence of a local clock, implying that the promoters of these genes have evolved to be directly responsive to circulating, systemic cues from the central nervous system.
Rhythms of snoRNA host genes
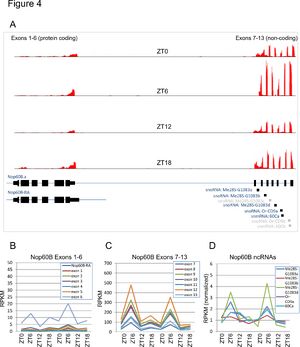
Non-coding, snoRNA host genes oscillate in the fly brain
We used Illumina sequencing to systematically profile RNA expression in fly heads over two consecutive days. We identified hundreds of cycling genes, including many non-coding RNAs (ncRNAs) that had not been identified in previous microarray studies. Most interestingly, an entire family of ncRNAs, the Uhg genes, oscillated over the course of 24 hours. These genes are hosts for snoRNA expression -- i.e., their exons are spliced together, and their introns are further processed to generate mature snoRNAs.
Since snoRNAs are involved in ribosomal maturation, this observation raises the possibility that these daily oscillations directly influence the assembly and stability of the ribosome, one of the most fundamental processes of cell biology. Alternatively, these snoRNAs may exert their influence over mRNAs instead, thereby regulating the stability or translation of circadian target genes. Consistent with this possibility, we found that snoRNAs can target several known clock genes in the fly, such as Takeout (to).

Hughes Lab
Pulmonary and Critical Care Medicine
Washington University School of Medicine
