BioMicroCenter:RNAseq
| HOME -- | SEQUENCING -- | LIBRARY PREP -- | HIGH-THROUGHPUT -- | COMPUTING -- | DATA MANAGEMENT -- | OTHER TECHNOLOGY |
The BioMicro Center supports several methods for RNAseq. The choice of method is highly dependent on the amount of input RNA and the quality of the input RNA. At this time, we only accept eukaryotic samples for RNAseq but we are willing to discuss exploring prokaryotic method development.
RNAseq Methods
| Amount of RNA | Quality of RNA | Method Recommended |
|---|---|---|
| >100ng | RIN:9.0 | Illumina TruSeq |
| >1ug | RIN:5.0 | Epicenter RiboZero |
| 10pg-100ng | RIN:9.0 | Clontech SMARTer Low Input |
| 1ng-1ug | RIN:5.0 | NuGEN Ovation RNA-Seq System V2 |
|
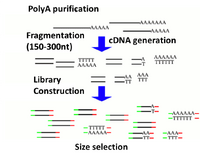
Illumina's TruSeq chemistry is the primary RNAseq method used in the BioMicro Center. This chemistry uses polyT beads to isolate the mRNA from the rRNA and tRNA. The use of these beads requires that the RNA be of very high quality or only the 3' end of transcripts will be isolated. Purified mRNA is then fragmented with metal and random priming is used to convert the sample to cDNA. Once double-stranded cDNA is generated, the sample is transferred to the SPRIworks for the remainder of sample preparation. |
 |
 |
For samples with degraded RNA or samples where you are interested in looking at non-polyA RNAs, the BioMicro Center utilizes the Epicenter ribominus kit. This kit uses *check me* magnetic beads coupled to rRNA and tRNA sequences to remove these sequences from the solution. The remaining mRNA fragments can then be converted in to cDNA. Once double-stranded cDNA is generated, the sample is transferred to the SPRIworks for the remainder of sample preparation.
|
For samples with less then 100ng of input, the BioMicro Center utilizes the Clontech SMARTer system. This system differs from the TruSeq chemisry in that it begins with cDNA generation using polyT priming and proprietary chemistry. The use of polyT priming requires the RNA to be of high quality. Full length double-stranded cDNAs are generated and amplified by PCR. These cDNAs are then fragmented and transferred to the SPRIworks for the remainder of sample preparation. Data from this system is of similar quality to samples created with Illumina TruSeq chemistry. |
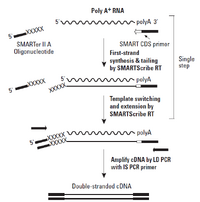 Image from Clonetech |
|
For samples with less then 100ng of input and restricted input amounts, our kit of choice is the NuGEN Ovation. This kit utilizes non-random nonamers designed to not amplify ribosomal RNA to create double stranded cDNA fragments. The fragmented cDNA is then transferred to the SPRIworks for the remainder of sample preparation. The downside of this kit is that the non-random nonamers cannot amplify all areas of the genome and certain portions of genes are often lost. Still, for many samples, this kit is the only way to generate RNAseq libraries. |
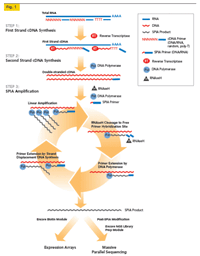 Image from NuGen. |
Additional Chemistries Available in the BioMicro Center
Strand Specific Sequencing
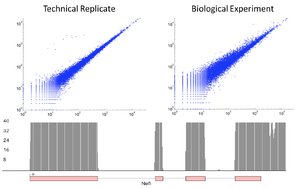
For some applications of RNAseq, such as splice choice determination, having a precise knowledge of the insert size is critical. While the SPRIworks does provide some size selection (typically restricting fragments to between 150 and 350bp), this can be too wide for some methodologies. In these cases, after libraries are amplified, they can be run on the Sage BluePippin (either singly or pooled). Here the size distribution can be much tighter, with most of the DNA fragments being within a 50nt range.
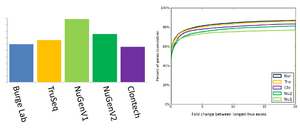
Comparison of the RNAseq methods
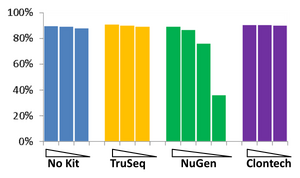
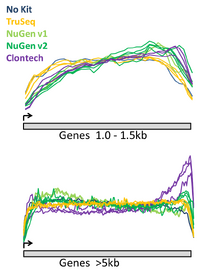
The BioMicro Center has done testing in head-to-head competitions of the TruSeq, NuGEN v2 and Clontech kits. These data were presented at AGBT 2012 as part of a poster. The authors were: Avanti Shrikumar, Zachary Banks, Manlin Luo, Ryan Sinapius, Paola Favaretto, Jessica Hurt, Chris Burge, and Stuart S. Levine. Selections of the poster are shown below:
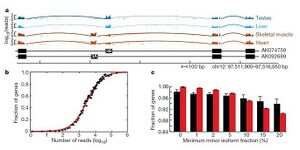
Choosing a read length and read depth
RNAseq References
A few notable references. Please feel free to add more:
<biblio>
- Paper1 pmid=18978772
- Paper2 pmid=19015660
- Paper3 pmid=18550803
- Paper4 pmid=20711195
<\biblio>