BIOL368/F14:Nicole Anguiano Week 6
From OpenWetWare
Jump to navigationJump to search
Star Biochem DNA Glycosylase Exercise
Methods and Results
- First, I downloaded Star BioChem, and began following the instructions from the DNA Glycosylase Exercise worksheet.
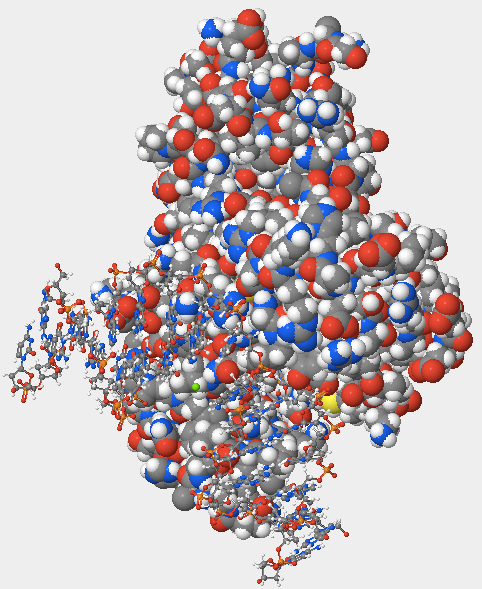
- I selected the “DNA glycosylase hOGG1 w/ DNA – H. sapiens (1EBM)” protein from the Samples > Select from Samples menu. I changed the size of the atoms from 20 to 100 to view the protein as a space-filling model.
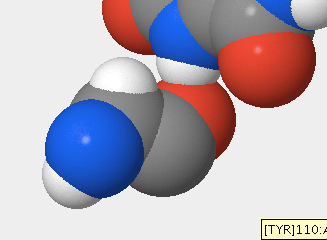
- Next, I pointed to a sulfur atom in the hOGG1 structure to obtain information about it.

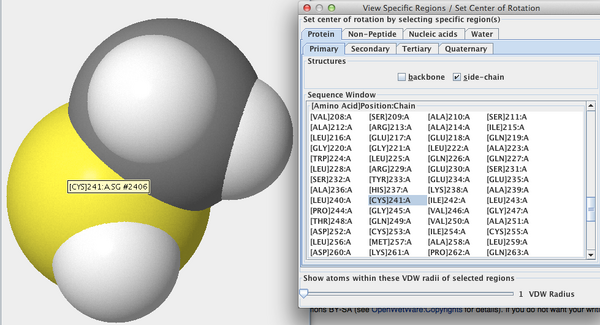
- I selected View > View Specific Regions / Set Center of Rotation, then selected the amino acid in Figure 2. I moved the VDW Radius slider to 1 and unchecked the side-chain box. I observed that the sulfur atom disappeared. I checked the side-chain box and unchecked the backbone box. The sulfur atom remained. From there, I deduced that the Sulfur atom is a part of the side-chain.
- I reset the structure by selecting Reset > Reset structure in the top menu, then scrolled down and found amino acids 105-117.
| Number | Amino Acid |
|---|---|
| 105 | Threonine (Thr) |
| 106 | Leucine (Leu) |
| 107 | Alanine (Ala) |
| 108 | Glutamine (Gln) |
| 109 | Leucine (Leu) |
| 110 | Tyrosine (Tyr) |
| 111 | Histidine (His) |
| 112 | Histidine (His) |
| 113 | Tryptophan (Trp) |
| 114 | Glycine (Gly) |
| 115 | Serine (Ser) |
| 116 | Valine (Val) |
| 117 | Aspartic Acid (Asp) |
- Table 1: The identity of amino acids number 105 to 117.
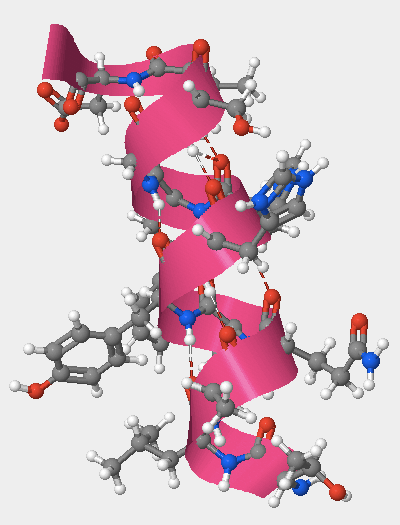
- I clicked on the secondary tab and selected "all" to view all of the different structures (helices, coils, and sheets). I moved the Structures Size slider to the right to make them larger. I selected View > View Specific Regions / Set Center of Rotation, and selected amino acids 105 to 117 by clicking on amino acid 105, holding Shift, then clicking on amino acid 117. I moved the VDM Radius slider to 1 so that I could just see the selected amino acids.
- I reset the structure by selecting Reset > Reset structure in the top menu. I selected the Tertiary tab and moved the atom slider to the left to make all the atoms as small as possible. I then selected the negatively charged/acidic radio button. I moved the Atoms Size slider to the right to make the acidic amino acids as large as possible.
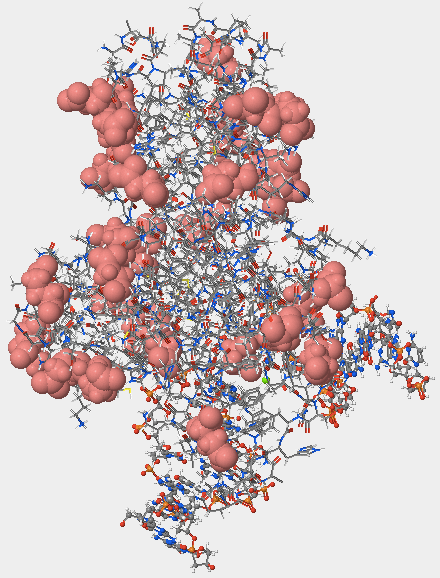
- I reset the structure by selecting Reset > Reset structure in the top menu. I selected the Nucleic Acids tab and moved the Atoms Size slider to 35. I then selected the Protein > Primary tab and moved the Atoms Size slider to 0 and the Bonds Translucency slider to 95. I selected the Nucleic Acids tab again and unchecked the phosphates checkbox and the sugar atoms checkbox. I moved the Atoms Size slider to 70. I clicked on the Non-Peptide tab and moved the Atoms Size slider to 100 so that I could fully observe the oxidized guanine.
- Without resetting the structure, I selected the Protein > Secondary tab and selected "all". Next, I selected the amino acids within helix 1 by clicking on its first amino acid, holding shift, and clicking the last amino acid in the helix. I increased the Secondary Structures Size slider to 100. I repeated the selection process with the amino acids for helix 16. I moved back to the Protein > Primary tab and selected the amino acids that corresponded to Helix 1, and unchecked backbone. I moved the Atoms Size slider to 100. I selected the amino acids in helix 16 to apply the sizing change to them as well.
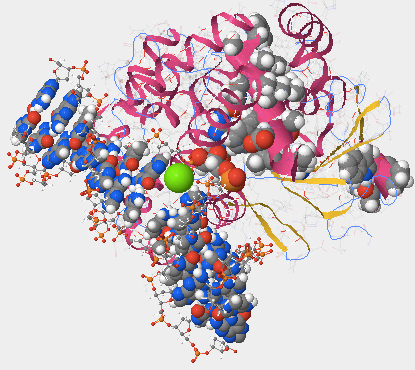
- In the top menu, I clicked on Samples > Select from Samples and selected “DNA glycosylase hOGG1 w/ DNA containing oxoG - H. sapiens (1YQR)”. In the Protein > Primary tab, I moved the Atoms Slize slider to the left to 0 and the Bonds Translucency Slider to 95. I clicked on oxidized guanine and moved the Atoms Size slider to 100. Then I went to the Protein > Primary tab and selected glycine 42, and moved the Atom Size slider to 100.
- I repeated the steps above with “DNA glycosylase hOGG1 w/ DNA containing G - H. sapiens (1YQR)”, only instead of increasing the size of the oxidized guanine, I increased the side of Guanine 23.
Questions
- The hOGG1 structure contains both DNA and protein. Can you differentiate between the DNA and protein components? How did you distinguish the DNA from the protein?
- It is possible to differentiate between the DNA and protein components. The hOGG1 structure's proteins are displayed in the space-filling form, but the DNA is still visible in the ball and stick model (Fig. 1). Therefore, the DNA appears much smaller and thinner than the protein, making it easily distinguishable from the protein.
- hOGG1 contains multiple sulfur atoms.
- Identify the name and sequence number of one of the amino acids in the structure that contains a sulfur atom.
- The sulfur atom in figure 2 is part of cysteine at position 241.
- Is the sulfur atom located in the backbone or in the side chain of the amino acid?
- The sulfur atom is located in the side chain of the amino acid (Fig. 3).
- Identify the name and sequence number of one of the amino acids in the structure that contains a sulfur atom.
- List the 13 amino acids numbered 105 through 117 in order.
- See table 1.
- Within a protein chain, amino acids form local structures called secondary structures.
- Are helices, sheets or coils present in hOGG1? Describe the color that represents the secondary structures you observe.
- Helices, sheets, and coils are all present in hOGG1. Helices are colored pink, sheets are colored yellow-orange, and coils are colored a darker blue-purple color.
- Amino acids 105 through 117 fold into one of the secondary structures. Which secondary structure do they fold into?
- Amino acids 105-107 fold into an alpha helix (Fig. 4).
- Are helices, sheets or coils present in hOGG1? Describe the color that represents the secondary structures you observe.
- Are the negatively charged amino acids located on the inside (buried) or outside (exposed) of this protein? What does that suggest about the cellular environment surrounding this protein, is it hydrophobic or hydrophilic? Explain your answer.
- The negatively charged amino acids are located on the outside (exposed) side of the protein (Fig. 5). This suggests that the cellular environment surrounding this protein is hydrophilic and made of a large amount of water. The amino acids on the outside will likely be exposed to the environment that is suitable to them. Negatively charged amino acids are hydrophilic, so the environment must be hydrophilic as well. If the environment was hydrophobic, the hydrophilic amino acids would repel the environment, and likely curve inwards so that they were not facing the outside.
- Now let’s explore how DNA glycosylase interacts with DNA to recognize the damaged DNA base within its sequence. DNA is composed of four bases: Adenine (A), Thymine (T), Cytosine (C) and Guanine (G). In this particular structure, the hOGG1 protein is bound to a segment of DNA that contains an oxidized guanine.
- How many DNA base pairs can you count within this double helix?
- I can count 13 DNA base pairs within the double helix.
- How many DNA bases are unpaired (not paired to its partner on the other strand)?
- There is one DNA base that is unpaired.
- Is the oxidized guanine base paired or unpaired? Describe the position of the oxidized guanine with respect to the hOGG1 protein and the double helix. What does this suggest about the mechanism that hOGG1 uses to identify damaged DNA bases?
- The oxidized guanine base is unpaired. The oxidized guanine is towards the center of the hOGG1 protein with the protein wrapping almost fully around it. It is also in the middle of the double helix. This suggest that hOGG1 uses the presence of oxidized guanine to determine whether or not a DNA base is damaged or not. The presence of it indicates the that base is damaged, and the absence of it indicates that the base is undamaged.
- How many DNA base pairs can you count within this double helix?
- Certain amino acids within hOGG1 form contacts with the DNA and are able to recognize if a guanine base has been damaged by oxidation. Where are you more likely to find the amino acids that recognize damaged guanine bases within hOGG1, in Helix 1 or Helix 16? Explain why.
- You are more likely to find the amino acids that recognize damaged guanine bases in Helix 16. Helix 16 is directly next to the oxidized guanine, and has a few atoms that are in direct contact with it (Fig. 6). Helix 1, on the other hand, is on the outside of the atom, quite far away from the oxidized guanine. The helix that is closest to the oxidized guanine will likely be the one responsible for identifying the damage, making it likely that helix 16 contains the amino acid that recognize the damaged guanine bases.
- hOGG1 is able to distinguish very efficiently between guanine and its oxidized counterpart, 8-oxoguanine. This represents a formidable task given that the oxidized nucleobase, 8-oxoguanine, differs by only two atoms from its normal counterpart, guanine (positions 7 and 8). The following structures illustrate hOGG1 bound to either an 8-oxoguanine or a guanine nucleobase: “1YQR” (8-oxoguanine) and “1YQK” (guanine). We will compare these two structures to understand how hOGG1 can effectively distinguish between 8-oxoguanine and guanine.
- Which atom in the base portion of 8-oxoguanine is directly contacting glycine 42? Draw this interaction.
- A hydrogen from 8-oxoguanine is directly contacting glycine 42 (Fig. 7). The hydrogen is on a nitrogen that is in-between two oxygens.
- What does the interaction between 8-oxoguanine and glycine 42 indicate about how hOGG1 differentiates between 8-oxoguanine and guanine?
- The regular guanine does not have a hydrogen on a nitrogen between two oxygens. There is only a single oxygen. If hOGG1 sees that there is a nitrogen between two oxygens, it knows to attach, but if the nitrogen is next to nitrogens and only one oxygen, then it knows not to attach.
- Compare the general location and orientation of guanine and glycine 42 in the “1YQK” structure with that of 8-oxoguanine and glycine 42 in the “1YQR” structure. How do the location and orientation of these two nucleobases and glycine 42 differ in these structures? Explain how this comparison adds to your understanding of hOGG1’s ability to discriminate between 8-oxoguanine and guanine.
- The location and orientation of guanine and glycine 42 in 1YQK and 1YQR are relatively similar, however the 8-oxoguanine is sticking almost directly out perpendicularly to the DNA molecule at a 90 degree angle with the backbone, while the base of the guanine 42 is at an angle. Due to the 8-oxoguanine sticking out at such an extreme perpendicularity to the backbone, the hOGG1 molecule may be able to more easily wrap around the defective guanine and know the change it. The larger size of the 8-oxoguanine may also help to facilitate this. The slight angle of the regular guanine likely discourages any bonds from forming between the protein and the DNA molecule.
- Which atom in the base portion of 8-oxoguanine is directly contacting glycine 42? Draw this interaction.
- Propose an experiment to test whether phenylalanine 114 plays a role in the detection and repair of 8-oxoguanine nucleobases.
- To test whether phenylalanine 114 plays a role in the detection and repair of 8-oxoguanine nucleobases, modify a set of DNA glycosylase MutM so that phenylalanine 114 is not present in them (either by removal or by changing the codon so that it codes for a different amino acid). Keep a second group of DNA glycosylase MutM in which phenylalanine is left intact. Then, see how capable the MutM protein is able to detect and repair 8-oxoguanine nucleobases, perhaps by measuring the number of 8-oxoguanine bases present at the end and comparing it with how many were present at the beginning. If phenylalanine 114 does play a role, then the modified MutM proteins should be ineffective at repairing the bases, while the intact MutM proteins will remain capable of doing so effectively. If phenylalanine 114 does not play a role, then the two groups should have minimal differences in the amount of 8-oxoguanine bases present in the DNA after a period of time.
Cn3D Exercise
Methods
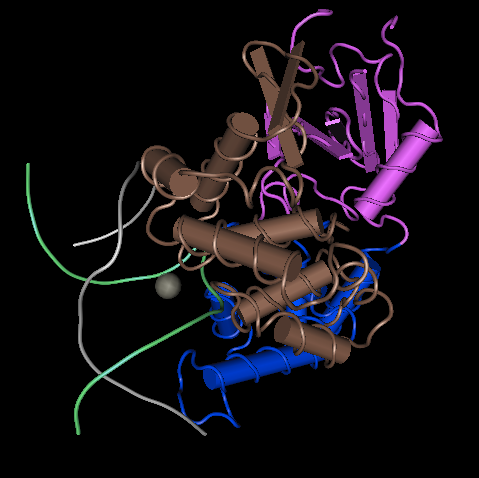
- First I downloaded the Cn3D file from NCBI. Next, I opened the file in Cn3D. I selected Style > Coloring Shortcuts > Domain to show the colors of the domains. Then I moved the protein structure until it was relatively similar to the position of Figure 6.
Questions
- Is this structure a tertiary structure or quaternary structure? Explain your reasoning.
- This structure is a quaternary structure. It contains the alpha helixes and beta sheets of tertiary structure, but contains several individual domains that together make up the quaternary structure.
- How many domains does DNA glycosylase have? Explain your reasoning.
- DNA glycosylase has four domains. There are four separate sections of the protein that, if separated form the protein, would be able to function on their own. This is especially evident when choosing the coloring scheme "Domain", which shows the four domains of the protein.
- Can you view the same things in the structure using this program as you did when you were using Star Biochem? Why or why not?
- It seems as though you can view the same things in Cn3D as you can in Star Biochem, though perhaps not in as much detail. Cn3D provides options to view the model in different formats (ball and stick, space filling), as in Star Biochem. It provides display of specific molecules through the sequence viewer. However, I'm not certain if it can display in as much detail as Star Biochem (ex, making one section bigger than the rest). While these options may be available, it is at least not extremely intuitive to figure out how to do it in Cn3D.
- Capture at least one screenshot that is a similar a view to one of your previous screenshots as possible and post it to your page.
- Which program do you prefer to use to view this structure? Why? You will choose to use one (or both) of these programs for your HIV Structure Project.
- I preferred using Star Biochem. The user interface was much easier to understand, and it was much simpler to figure out how to do the tasks that I wanted to do in Star Biochem than it was in Cn3D. While there are parts of CN3D that I preferred (namely the aesthetics and ease of zoom), I believe that the introductory tutorial of Star BioChem helped it to be much simpler to understand. The immediate presence of the options that can be used to edit the structure made using it much quicker.
Links
Nicole Anguiano
BIOL 368, Fall 2014
Assignment Links
- Week 1 Assignment
- Week 2 Assignment
- Week 3 Assignment
- Week 4 Assignment
- Week 5 Assignment
- Week 6 Assignment
- Week 7 Assignment
- Week 8 Assignment
- Week 9 Assignment
- Week 10 Assignment
- Week 11 Assignment
- Week 12 Assignment
- Week 13 Assignment
- Week 15 Assignment
Individual Journals
- Individual Journal Week 2
- Individual Journal Week 3
- Individual Journal Week 4
- Individual Journal Week 5
- Individual Journal Week 6
- Individual Journal Week 7
- Individual Journal Week 8
- Individual Journal Week 9
- Individual Journal Week 10
- Individual Journal Week 11
- Individual Journal Week 12
- Individual Journal Week 13
- Individual Journal Week 15