BIOL368/F14:Chloe Jones Week 6
Star Biochem DNA Glycosylase Exercise
Methods/Results
Methods were obtained from DNA Glycosylase Exercise
To start the exercise a molecular 3D viewer called StarBiochem was downloaded. In this exercise we observed the structure of hOGG1 which has a oxidized base ,changing the structure from guanine to 8-oxoguanine. To begin, I clicked the start button and then from the top menu I selected: select from samples within the Amino Acid/Proteins > Protein tab>“DNA glycosylase hOGG1 w/ DNA – H. sapiens (1EBM).”
- The hOGG1 structure contains both DNA and protein. Can you differentiate between the DNA and protein components? How did you distinguish the DNA from the protein?
- Yes. I differentiated between the DNA and protein components by looking at what atoms occupied the chains. I know if a nitrogen and oxygen were both being occupied by carbons that were adjacent to one another I would consider that to be a protein .For the DNA I looked for a sugar phosphate backbone and that was present in the ball and stick model which showed an alpha helical structure. Proteins are able to fold into multiple structures, so there was not a defined shape.
- hOGG1 contains multiple sulfur atoms .a.) Identify the name and sequence number of one of the amino acids in the structure that contains a sulfur atom. b.) Is the sulfur atom located in the backbone or in the side chain of the amino acid?
- a.) CYS:75:A SG #1138 Amino Acid: Cysteine position: 75
- b.) Side chain of the amino acid, able to recognize this by unchecking the backbone box.
- Methods
- b.) From the top menu I selected, View> View Specific Regions / Set Center of Rotation. I then selected the amino acid, Cysteine at position 75, which I had chosen previously. I moved the VDW Radius slider all the way to the left (1 Van der Waals radii), and then zoomed to my preference. I unchecked the side-chain box, and the backbone box to separately view the backbone and side chains of the amino acids.
- Methods
- Next, I explored the primary structure of the hOGG1 protein .The hOGG1 protein consists of 325 amino acids. List the 13 amino acids numbered 105 through 117 in order.
- hOGG1 proteins: Threonine(105), Leucine(106), Alanine(107), Glutamine(108), Leucine(109), Tyrosine(110), Histidine(111), Histidine(112), Tryptophan(113), Glycine(114), Serine(115), Valine(116), Aspartate(117)
- Methods
- Using the Sequence Window to the left in the Protein>Primary tab, the amino acids 105-117 were able to be located. The reference page then allowed me to transcribe the full name from the three letter codes that were presented.
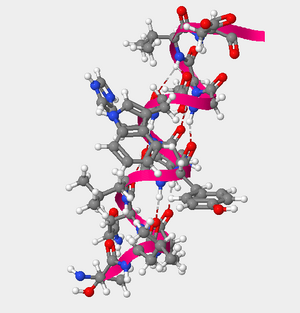
Figure 1. The helcial structure of amino acids 105 through 117 of the hOGG1 protein.
- Using the Sequence Window to the left in the Protein>Primary tab, the amino acids 105-117 were able to be located. The reference page then allowed me to transcribe the full name from the three letter codes that were presented.
- Methods
- hOGG1 proteins: Threonine(105), Leucine(106), Alanine(107), Glutamine(108), Leucine(109), Tyrosine(110), Histidine(111), Histidine(112), Tryptophan(113), Glycine(114), Serine(115), Valine(116), Aspartate(117)
- Explore the secondary structures found in hOGG1. a.) Are helices, sheets or coils present in hOGG1? Describe the color that represents the secondary structures you observe. b.) Amino acids 105 through 117 fold into one of the secondary structures. Which secondary structure do they fold into?
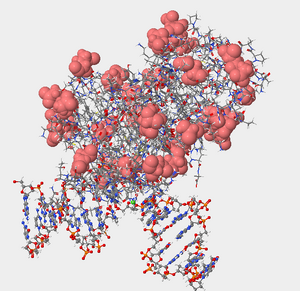
Figure 2. Negatively charged amino acids are hydrophilic and will be located on the exterior surface of the protein exposed to the environment. Denoted in the picture by the salmon color. 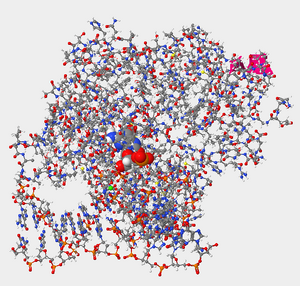
Figure 3. Helix 1 within the protein hOGG1 in relation to damaged guanine bases (enlarged in photo). 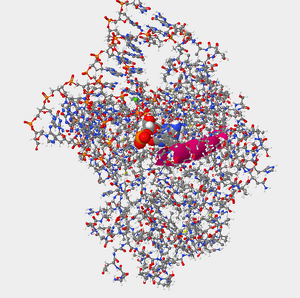
Figure 4. Helix 16 within the protein hOGG1 in relation to damaged guanine bases (enlarged in photo). - a.) Yes.Alpha Helices=Fuschia/Pink, Beta Sheets=Yellow, Coils= Blue
- Methods
- I clicked on the secondary tab, and then I selected the box that was beside the structure of interest (i.e. beta sheets), I used the slider to zoom if necessary.
- Methods
- b.) Answer: Helices
- Methods
- From the secondary tab I selected the all bottom which made the amino acids visible to be selected. I then selected the amino acids of interest which were 105-117(clicked 105 then held down shift and clicked 117). I slid the VDW radius all the way to the left which made only the selected amino acids viewable.
- Methods
- a.) Yes.Alpha Helices=Fuschia/Pink, Beta Sheets=Yellow, Coils= Blue
- Negatively charged amino acids are hydrophilic (Reference page). Are the negatively charged amino acids located on the inside (buried) or outside (exposed) of this protein? What does that suggest about the cellular environment surrounding this protein, is it hydrophobic or hydrophilic? Explain your answer.
- Negatively charged amino acids are hydrophilic so they are on the outside exposed because they are essentially “water loving.” The cellular environment surrounding the protein as well must by hydrophilic so there can be interaction between the polar charged and non-polar charged amino acids.
- Methods
- I reset the structure. I then clicked on the tertiary tab, and moved the bar to the left decreasing the size of the atoms. I then clicked the charged/acidic button, and increased the atom size to make the negatively charged/acidic amino acids more visible
- Methods
- Negatively charged amino acids are hydrophilic so they are on the outside exposed because they are essentially “water loving.” The cellular environment surrounding the protein as well must by hydrophilic so there can be interaction between the polar charged and non-polar charged amino acids.
- In this particular structure, the hOGG1 protein is bound to a segment of DNA that contains an oxidized guanine. First, let’s make the DNA segment more visible. a) How many DNA base pairs can you count within this double helix? b) How many DNA bases are unpaired (not paired to its partner on the other strand)? c) Is the oxidized guanine base paired or unpaired? Describe the position of the oxidized guanine with respect to the hOGG1 protein and the double helix. What does this suggest about the mechanism that hOGG1 uses to identify damaged DNA bases?
- Method: to make DNA segment more visible. In the Nucleic Acid tab, I moved the Atom size slider to right by 35% and in the Protein>Primary tab I moved the Atoms Size slider completely to the left and the Bonds Translucency slider to the right by 95%.
- a & b) Answer 6a: 14 Answer 6b: : 2 unpaired bases
- Methods
- I clicked on the Nucleic Acid tab, and unchecked the phosphates and sugars boxes. To count the base pair more easily I moved the Atom Size slider to 70%, and l left the phosphates and sugar nucleotide components intact.
- Methods
- c.) Answer: Unpaired. The oxidized guanine is located between the protein and the start of the two DNA helices. This suggest that the hOGG1 proteins uses orientation of the amino acid to determine if it does not properly fit within the molecule.
- Methods
- I clicked on the Non-Peptide tab and moved the atom size slider completely to the right 100% to increase the oxidized guanine ([8OG]25).
- Methods
- Certain amino acids within hOGG1 form contacts with the DNA and are able to recognize if a guanine base has been damaged by oxidation. Where are you more likely to find the amino acids that recognize damaged guanine bases within hOGG1, in Helix 1 or Helix 16? Explain why.
- Helix 1 relative to the oxidized guanine is on the outskirts, so Helix 16 would be a more appropriate option. Helix 16 is composed of polar side chains which were referenced in the reference page and thus would be more likely to interact with the oxidized guanine. Overall, it would be more effective because of the location relative to the oxidized guanine and the amino acids that make up Helix 16.
- Methods
- I clicked the All button in the Protein> Secondary tab. I then selected the amino acids which were in Helix 1 which contained 5 amino acids I then did the same for Helix 16 which contained 14 amino acids. I also noted where the helices were in regards to oxidized guanine. I then clicked the Protein >primary tab too see if any of the side chains of Helix 1 or Helix 16 were in contact with ([8OG]25). Selecting for Helix 1 and then for Helix 16 individually, I unchecked the backbone and then enlarged the atoms to see any interactions.
- Methods
- Helix 1 relative to the oxidized guanine is on the outskirts, so Helix 16 would be a more appropriate option. Helix 16 is composed of polar side chains which were referenced in the reference page and thus would be more likely to interact with the oxidized guanine. Overall, it would be more effective because of the location relative to the oxidized guanine and the amino acids that make up Helix 16.
- a) Which atom in the base portion of 8-oxoguanine is directly contacting glycine 42? b) What does the interaction between 8-oxoguanine and glycine 42 indicate about how hOGG1 differentiates between 8-oxoguanine and guanine? c.) Compare the general location and orientation of guanine and glycine 42 in the “1YQK” structure with that of 8-oxoguanine and glycine 42 in the “1YQR” structure. How do the location and orientation of these two nucleobases and glycine 42 differ in these structures? Explain how this comparison adds to your understanding of hOGG1’s ability to discriminate between 8-oxoguanine and guanine
- 8a.) The hydrogen located in 8-oxoguanine 8b.) In the oxidized version glycine 42 and 8-oxoguanine share an interaction where the hydrogen of oxoguanine is connected with the oxygen of glycine 42. Glycine 42 attaches to the H in oxoaguanine whereas a nitrogen is present in guanine. A bond cannot form between the oxygen of the glycine 42 and the nitrogen of the guanine.
- Methods
- I clicked on Select from sample as the top, then within the Amino Acid/Proteins>Proteins tab I selected “DNA glycosylase hOGG1 w/ DNA containing oxoG - H. sapiens (1YQR)” which is the hOGG1 bound to a 8-oxoguanine. In the Protein> Primary I moved the Atom size completely to the left (0%) and the Bonds Translucency slider completely to the right (100%). Then I clicked on the Nucleic Acids tab and increased the size of the oxidized guanine with the Atoms Size slider. I then went back to the Protein>primary tab and clicked glycine 42 and increased its size as well. I then repeated the steps for “DNA glycosylase hOGG1 w/ DNA containing G - H. sapiens (1YQK)” selecting and enlarging the sequences to see the interactions.
- Methods
- c.) In the “1YQR” structure there is not a direct bond but rather an orientation of the oxygen from the glycine 42 to towards the hydrogen of the guanine. However, in “1YQK” the glycine 42 is directly bonded to the 8-oxoguanine. Based upon the orientation when an oxidized guanine is present vs. when a regular guanine is present allows us to see that hOGG1’s ability to discriminate depends readily on the conformational changes that the amino acids in encounter in regards to the guanine.
- Methods
- I reset the structure of “YQR”, by selecting reset>reset structure located in the top menu. I then moved the Atoms Size slider completely to the left and the Bonds Translucency slider to completely to the right (95%) to reduce the appearance of amino acis. Next, I clicked on the Nucleic Acids tab and selected the guanine at position 23 and enlarged it by moving the Atoms Size slider to the right. I then went back to the Protein>primary tab and clicked glycine 42 and moved the Atoms Size slider to the right.
- Methods
- 8a.) The hydrogen located in 8-oxoguanine 8b.) In the oxidized version glycine 42 and 8-oxoguanine share an interaction where the hydrogen of oxoguanine is connected with the oxygen of glycine 42. Glycine 42 attaches to the H in oxoaguanine whereas a nitrogen is present in guanine. A bond cannot form between the oxygen of the glycine 42 and the nitrogen of the guanine.
- Propose an experiment to test whether phenylalanine 114 plays a role in the detection and repair of 8-oxoguanine nucleobases
- Experiment:First, I would run PCR on the sample in order to get multiple copies of the DNA in order to test if phenylalanine 114 plays a role in the detection of an oxidized guanine. I would then perform a western blot with phenylalanine 114 and the segment of DNA of interest. After the blot is on a membrane I would then use immunofluorescence staining with an antibody that recognizes phenylalanine 114 to see if it intercalated at the point where the oxidized guanine was present. To see if phenylalanine 114 helps with repair I would run an experiment with a DNA segment that had no phenylalanine 114 present, and a DNA segment with phenylalanine 114 present. After that I would observe the confirmation and the orientation of the amino acids surrounding the molecule to see if it went back to what is normally observed in a guanine when phenylalanine 114 was present.
Cn3D
Methods/Results
From the NCBI Structure database I downloaded Cn3D. In the search engine I typed in “1ebm” which then allowed me access to view the DNA Glycosylase in the Cn3D program. Using the various menu options I was able to color the structure based on the different guidelines that were offered.
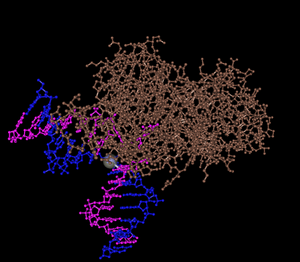
- Is this structure a tertiary structure or quaternary structure? Explain your reasoning.
- The structure that is shown in a quaternary structure because it is a multi subunit complex that is made out of more than one protein molecule.
- How many domains does DNA glycosylase have? Explain your reasoning.
- DNA glycosylase has three domains which were observed in the program because they were able to be differentiated by color.
- Can you view the same things in the structure using this program as you did when you were using Star Biochem? Why or why not?
- The overall ball and socket structure is the same for both programs, but the overall specificity was better in Star Biochem. Star Biochem allowed for certain nucleotides to be highlighted and enlarged to further examine which was not an available feature in the Cn3D program. The Star Biochem had more features that allowed the viewer to become more acquainted with the molecule and its amino acids individually. However, Cn3D was easier in examining the bigger picture because of how it allowed bigger subunits to be color coordinated. I felt as though Cn3D had other factors that could be beneficial such as hydrophobicity, pH, and charge but they were not as useful in this present experiment.
- Capture at least one screenshot that is a similar a view to one of your previous screenshots as possible and post it to your page.
- See Figure 5 (similar to Figure 2)
- Which program do you prefer to use to view this structure? Why? You will choose to use one (or both) of these programs for your HIV Structure Project.
- Both programs for the most part were easy to use and added some insight to my knowledge of the protein, but I think I preferred Star Biochem because it allowed better observance of what was occurring individually amongst the amino acids. I liked that the atoms could be zoomed in or made invisible at your discretion when studying the protein. However, I will probably use both for my HIV structure Project because I liked the color-coding tab that the Cn3D program had to offer because it allowed for easy differentiation.
Electronic Lab Notebook
- Chloe Jones Week 2
- Chloe Jones Week 3
- Chloe Jones Week 4
- Chloe Jones Week 5
- Chloe Jones Week 6
- Chloe Jones Week 7
- Chloe Jones Week 8
- Chloe Jones Week 9
- Chloe Jones Week 10
- Chloe Jones Week 11
- Chloe Jones Week 12
- Chloe Jones Week 13
- Chloe Jones Week 15
Weekly Assignments
- Week 1 Assignment
- Week 2 Assignment
- Week 3 Assignment
- Week 4 Assignment
- Week 5 Assignment
- Week 6 Assignment
- Week 7 Assignment
- Week 8 Assignment
- Week 9 Assignment
- Week 10 Assignment
- Week 11 Assignment
- Week 12 Assignment
- Week 13 Assignment
- Week 15 Assignment
Class Journals
- Class Journal Week 1
- Class Journal Week 2
- Class Journal Week 3
- Class Journal Week 4
- Class Journal Week 5
- Class Journal Week 6
- Class Journal Week 7
- Class Journal Week 8
- Class Journal Week 9
- Class Journal Week 10
- Class Journal Week 11
- Class Journal Week 12
- Class Journal Week 13
- Class Journal Week 15
Chloe Jones 03:46, 15 October 2014 (EDT)Chloe Jones