Amanda N. Wavrin Week 8
From OpenWetWare
Jump to navigationJump to search
Analyzing Protein Structure
- Using Bioinformatics for Dummies
Retrieving Protein Sequences
- Used the ExPASy server to search DUTPase coli, 105 results appeared
- Then searched gp120 and decided to use envelope glycoprotein gp120 with accession number A0A897
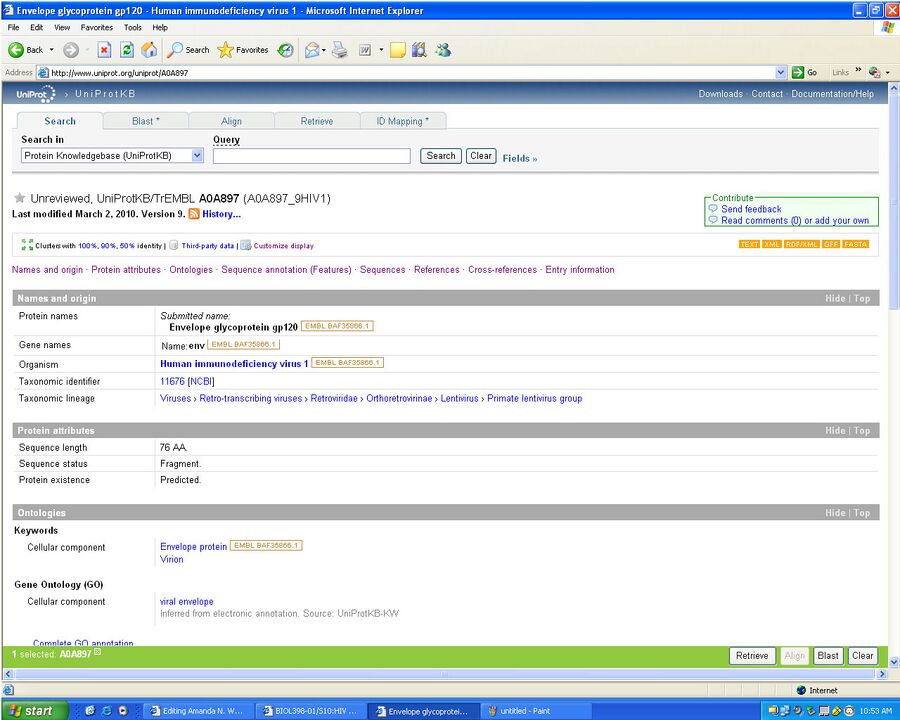
- We then retrieved the FASTS Format of the gp120 sequence
- >tr|A0A897|A0A897_9HIV1 Envelope glycoprotein gp120 (Fragment) OS=Human immunodeficiency virus 1 GN=env PE=4 SV=1
NLTDNAKTIIVHLNESVEINCTRPFNNTRTSXRIGPGQVFYRTGDITGSIRRAYCEINGT KWNKVLXQVTEKLXEH
- We then conducted an advanced search, starting back at the Swiss-Prot home page
Reading a Swiss-Prot Entry
- we began at the UniprotKB/Swiss-Prot page of the expasy server
- Then searched A0A987 for our gp120 protein
- There a section containing the name and the origin of the protein
- There is a refrences section wtih all of the bibliographical refrences used
- There is a cross-refrences section which contains refrences to other aticles containing iformation about this specific gp120 protein
- For the gp120 protein there is no extra comments section
- There is also a key word section containing keywords, envelope proten and virion
- There is a features section containing experimental information
ORFing your DNA sequence
- Used the NCBI ORF Finder site
- Chose to use subject 2, visit 4, clone 2 (S2V4-2)
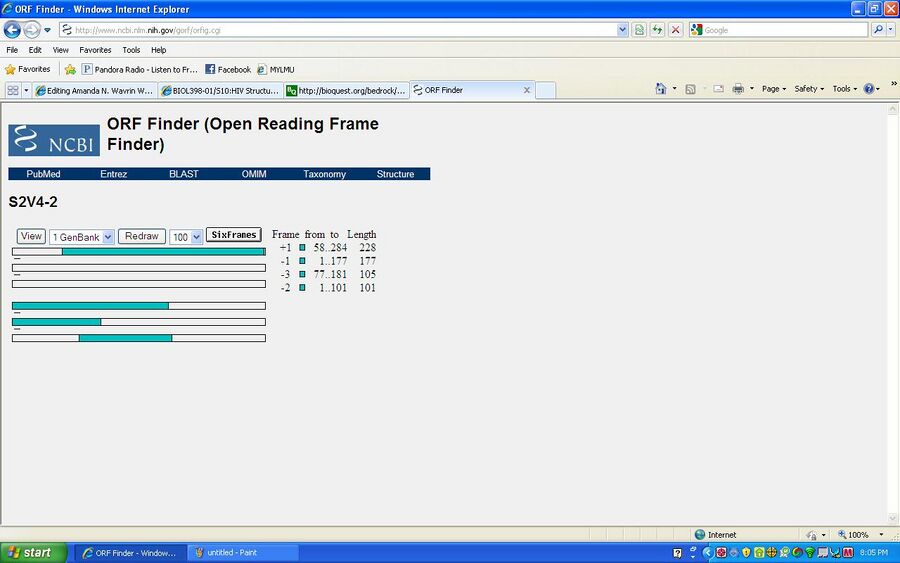
- By clicking on the on the rectangles in the display, wou could examine the ORFs mroe closely
Working with a Single Protein sequence
Predicting the main physico-chemical properties of a protein
- We used the primary structure analysis section of the ExPASy tools page
- We then entered the accession number A0A987 from Swiss-Prot into the ProtParam link site
- ProtParam generated the parameters of the entire gp120 sequence
- The results included the number of amino acids in the sequence, the molecular weight, the total number of positively charged residues, the total number of negatively charged residues, and more.
Digesting a protein in a computer
- We pasted the gp120 seguence into the ExPasy website to cut the protein in specific ways
Looking for transmembrane segmentsing
- We entered the accession number A0A987 into the ExPASy ProtScale site and submited it with the full range of analysis
- Then retrieved the image in GIF format
Interpreting ProtScale results
- We used a piece of paper to help locate the strongest peaks of the graph
- Using this technique we identified four transmemrane regions
Running TMHMM
- We ran TMHMM by pasting the FASTA sequence of our gp120 protein into the search box
- This generated the TMHMM results
Looking for PROSITE patterns
- Went to the ExPASy ScanProsite page
- Pasted the accession numbber A0A897 into the proteins to be scanned section and started the scan
Finding domains with InterProScan
- Went to the InterProScan sequence search page
- Pasted the gp120 sequence that we have been working with into the search box
- We removed some of the larger databases from the search to speed up the search
- After conducting the search, we examined the results and interpreted them by following the guide in Bioinformatics for Dummies
Finding domains with the CD server
- CD= Conserved Domain
- For this we used the NCBI Conserved Domains Search site
- Pasted the FASTA Format of the gp120 protein into the search box
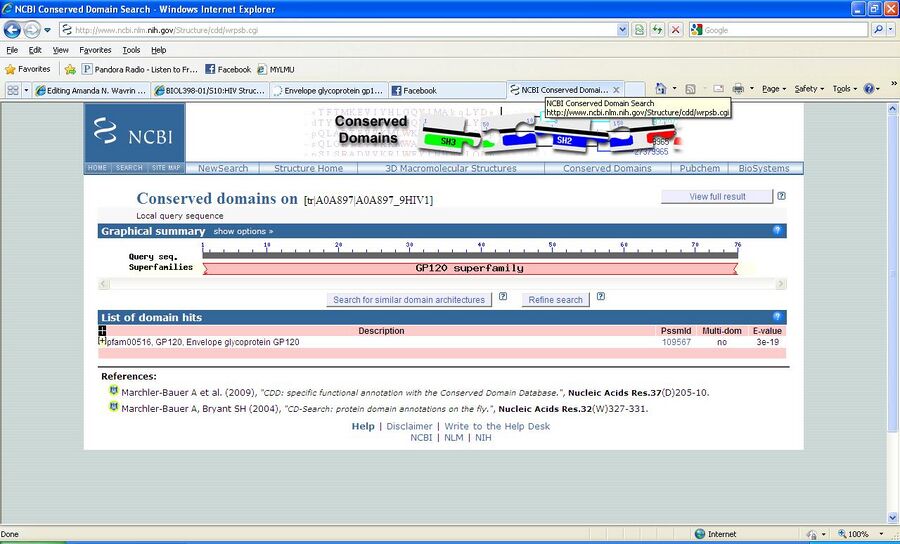
- We then interpreted these results with help from the Bioinformatics for Dummies book
Finding domains with Motif Scan
- To do this we used the Motif Scan website
- Pasted the FASTA sequence into the Input box
- Selected PROSITE and then conducted a search
- The results included a matches map, a list of matches, and a matches details section
Useful Links
- My User Page
- Open wetware Help Page
- Yahoo Home Page
- Class Journal Week 2
- Class Journal Week 3
- Class Journal Week 4
- Class Journal Week 5
- Class Journal Week 6
- Class Journal Week 7
- Class Journal Week 8
- Class Journal Week 9
- Class Journal Week 11
- Class Journal Week 12
- Class Journal Week 13
Weekly Journals
- Amanda N. Wavrin Week 2
- Amanda N. Wavrin Week 3
- Amanda N. Wavrin Week 4
- Amanda N. Wavrin Week 5
- Amanda N. Wavrin Week 6
- Amanda N. Wavrin Week 7
- Amanda N. Wavrin Week 8
- Amanda N. Wavrin Week 9
- Amanda N. Wavrin Week 11
- Amanda N. Wavrin Week 12
- Amanda N. Wavrin Week 13