Amanda N. Wavrin Week 4
From OpenWetWare
Jump to navigationJump to search
Activity 1, Part 2: GenBank
- Accesion number of chosen sequence: AF089135
- This seqence was from subject 3
- The definition tells us that the sequence was from S3V5-4
- Example FASTA sequences
Activity 1, Part 3: Introduction to the Biology Workbench
- Set up an account for Biology Workbench!
- Saved my FASTA seguence examples into Biology Workbench
- Used the ClustalW tool in Biology Workbench to perfroms multiple sequence alignment on the FASTA sequences
Activity 2, Part 1: Looking at the sources of HIV across subjects
- Uploaded all of the visit 1 nucleic acid sequences
- Generated a multiple sequence alignment for 12 sequences (3 clones from each of 4 subjects)
- Subject -- Clone number
- 12 -- 2,3,4
- 13 -- 2,3,4
- 14 -- 4,5,6
- 15 -- 10,11,12
- Unrooted Tree
- Yes, the clones from each subject cluster together
- yes, some subject's clones show more diversity than others. The branches of subjects 12 and 13 are longer than the others, showing greater genetic distance.
- Subjects 15 and 14 cluster closer together
- Each of the clones from each subject cluster together. The closer clustering of subject 14 and 15 clones suggests that sequences 14 and 15 are more similar to eachother than to the other sequences. The long branches of subjects 12 and 13 suggest greater genetic distance from the other sequences.
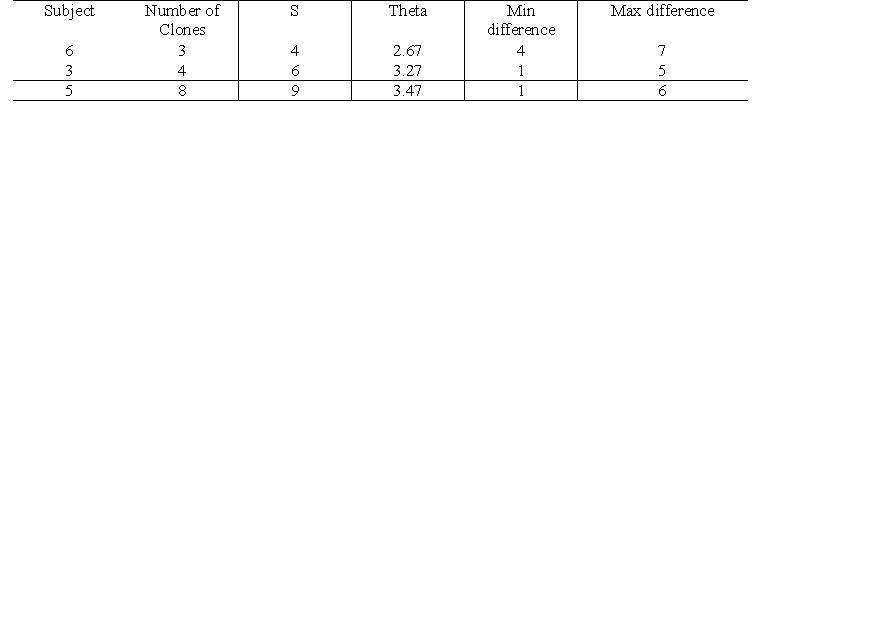
Activity 2, Part 2: Quantifying diversity within and between subjects
- Selected all the clones for subject 6, visit 1 and ran a multiple alignment
- Imported alignments and used the Clustdist tool to generate a distance matrix
- Selected all the clones for subject 3, visit 1 and ran a multiple alignment
- Imported alignments and used the Clustdist tool to generate a distance matrix
- Selected all the clones for subject 5, visit 1 and ran a multiple alignment
- Imported alignments and used the Clustdist tool to generate a distance matrix