Alex J. George Week 3
From OpenWetWare
Jump to navigationJump to search
Week 3: HIV Introduction
- My knowledge about HIV is definitely limited to what I hear in the news and media. I know HIV stands for the "Human Immunodeficiency Virus". Also, I know that you must be HIV + in order to contract AIDS. My understanding is that being exposed to the blood of an HIV + is enough to become HIV + yourself. Other than this basic information, I am not very knowledgeable about the biology of HIV.
- Questions concerning HIV:
- Is it possible to be born HIV + without your mother being HIV +?
- What is the biological affect of being HIV + on humans? What is physically/chemically changed in a human between HIV-/HIV+?
- Has HIV been attributed to any particular gene/chromosome? What is the most we know about the HIV virus?
PubMed Activity
- Pubmed's Menu is a little different than the text (There are no tabs in the newer version)
- Searching "dUTPase" turns up multiple results, but few define what it is. So, I clicked on the suggested search "human dUTPase" under the "Also Try" menu
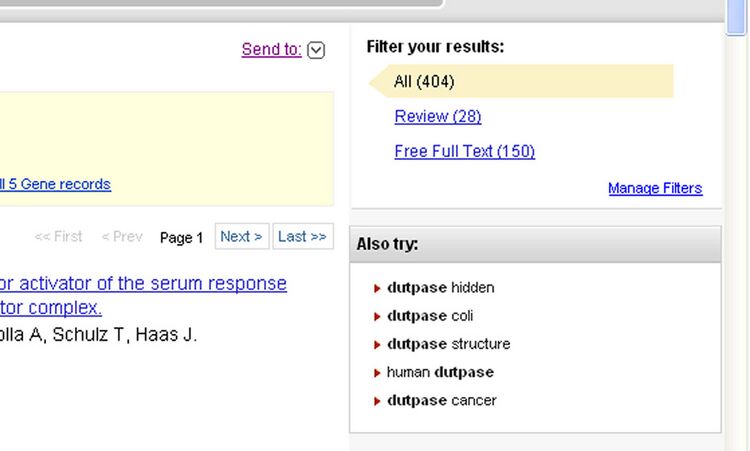
- This narrowed my search allowing me to discover the purpose of dUTPase on the first result, Kinetic properties and specificity of trimeric Plasmodium falciparum and human dUTPases.
HIV Article
- In order to find a useful HIV article on Pubmed, I searched "Human Immunodeficiency Virus" to search for articles with exactly that phrase. Since I only was interested in Full Text articles, I clicked on "Free Full Text" along the right side of the page.
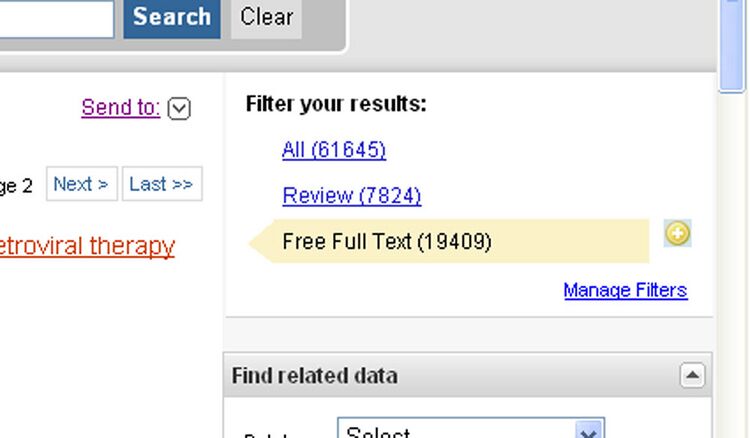
- I found a related article that defines the HIV virus by skimming the titles and abstracts. The article I found was the 24th result, Alternative paths in HIV-1 targeted human signal transduction pathways.
- In order to see the full text for this article, scroll down, click on "LinkOut- more resources", then select one of the links under the "Full Text Sources" Header:
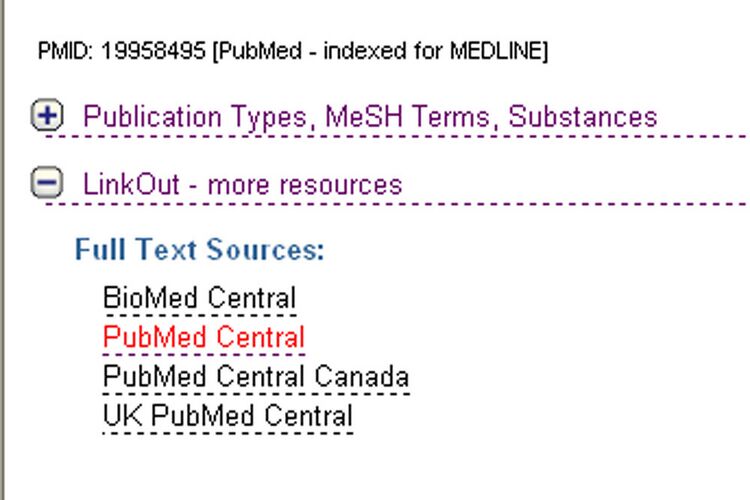
- The "Pubmed" link (highlighted above) takes you to this version of the full text: paths in HIV-1 targeted human signal transduction pathways.
Comparison to Google Scholar and ISI Web of Science
- The results for a Google Scholar search "Human Immunodeficiency Virus" seem to be more relevant than the results in Pubmed. The third result explains the expression of HIV in T Cells. The format of Google Scholar, however is worse than PubMed. The search results don't seem categorized and aren't as easy to distinguish.
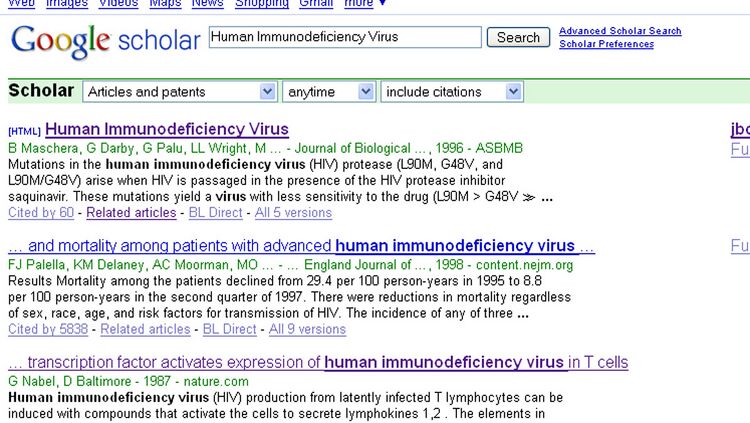
However, I do like the "Related articles" link under each result which makes it easier to find relevant information. Also, there seem to be more full text articles within Google Scholar. Overall, I like the results given by Google scholar, but it is much harder to navigate.
- The ISI Web of Knowledge Database is much more organized and makes much more sense to the user. I love the bar on the side to limit results. This provides a lot of options to limit or expand results. A simple "Human Immunodeficiency Virus" search under "Topic" can be limited to Review articles.
Overall, this is my favorite database. I feel like it is much easier to find what you are looking for.
Markham Article
- Since it's Publication, the article was cited 52 times.

Definitions
- Seroconverting- development of detectable specific antibodies to microorganisms in the blood serum as a result of infection or immunization
- Virology- the study of viruses and viral diseases
- Nested PCR- (Polymerase Chain Reaction)- technique to magnify and copy pieces of DNA; nested is intended to reduce contamination
- Peripheral- constituting the outer boundary
- CD4 T cell- recognizes antigens on the surface of a virus-infected cell and secretes lymphokines that stimulate B cells and killer T cells; AIDS knocks out these cells
- Sanger Chain Termination method- method of sequencing DNA using modified bases to stop replication
- pUC19- plasmid cloning vector in which DNA can be inserted
- Plasma viral load- measure of the severity of a viral infection, especially HIV
- Monophyletic- common descent from a single ancestor
- epidemiology- the study of factors that affect the health and illness of populations
Summary
Introduction
- High mutation/replication rates of HIV-1 lead to rapid adaptation
- In stable conditions, most viable ("fit") mutant will be most prevalent
- If unstable environment exists, either variable mutants are selected against or the dominant strain is selected against
- If the dominant strain is selected against, then the diversity of mutants increases too much, such that the immune system can't control it
- In HIV, the study looks at the forces against HIV that cause diversity and look at how the virus is adapting to forces
Limitations of Previous Work
- Didn't look at sequence patterns
- Subject pools were too small
- The McDonald study didn't follow subjects from seroconversion and had limited "time point" for analysis
Methods
- Study Population
- Took 15 participants who were known injection users
- Collected blood every 6 months and categorized participants as "Rapid progressors", "moderate progressors" and "non-progressors"
- Sequencing HIV-1 Genes
- Amplified 285-bp region of "env" gene from PBMC (peripheral blood mononuclear cells)
- Used primers to find distinct sequences
- Phylogenetic Trees
- Constructed using a computer program
- Indicates time when each strain was isolated and the number of identical replicated sampled
- Shows independent segregation of clones
- Correlation Analysis
- Analyzed correlation between Genetic Diversity vs. CD4 Cell count and Mutational Divergence vs. CD4 cell count
Figures
- Figure 1: shows the diversity, divergence, and CD4 cell count for each participant over time
- Figure 2: Shows "mean slope" of diversity and divergence of each progressor category and indicates significant differences
- Table 1: Summary of data on all subjects
- Figures 3 & 4: Phylogenetic trees of subjects 5,7,8,9, and 14. Shows evolution is not sustained along single branch
Results
- Main result: The more genetic diversity of the HIV-1 virus, the more rapid decline is in CD4 T-cell count
- CD4 decline was variable
- Sequence analysis performed on V3 domain (which is important for host-virus interaction)
- Diversity and divergence increased over time
- Diversity and divergence significantly negatively correlated to CD4 cell count
- The dS/dN ration for Rapid and Moderate progressors was .4, indicating an advantage for nonsynonymous changes
- For both divergence and diversity, rapid progressors differed significantly from nonprogressors
- Phylogenetic trees gave no evidence of predominance by single strain
Discussion
- Increase in diversity and divergence in HIV-1 variants led to CD4 decline
- Nonprogressor viral strains showed possible selection against amino acid change, whereas moderate/rapid progressors selected for amino acid change
- For subjects who contracted AIDS, their diversity and divergence continued to increase
- To control infection, the host cell must control it at an organismal level due to high diversity of virus mutants
- The importance of this work is that it reveals an more logical route to attacking HIV in patients who have already been diagnosed with the virus.
- Results were consistent with that of M.A. Nowak, but differed with two other recent studies conducted by McDonald and Wolinsky et al.
- McDonald's study showed that rapid progressors had less diversity than nonprogressors
- Wolinsky also observed less diversity in 2 subjects with the most rapidly declining CD4 T-cell count
Individual Journals
- No Week 1 Journal
- Alex J. George Week 2
- Alex J. George Week 3
- Alex J. George Week 4
- Alex J. George Week 5
- Alex J. George Week 6
- Alex J. George Week 7
- Alex J. George Week 8
- Alex J. George Week 9
- Alex J. George Week 10
- Alex J. George Week 11
- Alex J. George Week 12
- Alex J. George Week 13