Lidstrom:Buffers: Difference between revisions
| Line 102: | Line 102: | ||
* Michael Konopka's words: "What assays are you trying to measure? As for the buffering capacity, that really shouldn't matter since the pertinent components for the buffer (mono- and di-basic phosphate) are the same. The issue is if the potassium or sodium ion will form a precipitate which you don't want around if mixing with other solutions (or do want). A classic example is potassium will precipitate SDS while sodium is soluble. That's why in minipreps one adds potassium acetate/acetic acid after using SDS/NaOH to lyse the cells/dissolve lipids & proteins. The proteins, lipids, and chromosomal DNA is then trapped in the precipitate (plasmid DNA still in solution)." | * Michael Konopka's words: "What assays are you trying to measure? As for the buffering capacity, that really shouldn't matter since the pertinent components for the buffer (mono- and di-basic phosphate) are the same. The issue is if the potassium or sodium ion will form a precipitate which you don't want around if mixing with other solutions (or do want). A classic example is potassium will precipitate SDS while sodium is soluble. That's why in minipreps one adds potassium acetate/acetic acid after using SDS/NaOH to lyse the cells/dissolve lipids & proteins. The proteins, lipids, and chromosomal DNA is then trapped in the precipitate (plasmid DNA still in solution)." | ||
== Notes == | == Notes specific to buffers == | ||
=== phosphate buffer === | |||
* Phosphates, for example, form insoluble salts with bivalent metals and precipitate. Phosphate buffered salt solution (PBS) is never | |||
autoclaved with Ca2+ or Mg2+ for this reason. Good buffers, such as PIPES, TES, HEPES and CAPS have very low metal-binding | |||
constants and are therefore particularly suited to investigate metal-dependent enzymes (Good & Izawa 1972, Blanchard | |||
1984). [http://www.applichem.com/fileadmin/Broschueren/BioBuffer.pdf Applichem] | |||
Revision as of 23:50, 20 January 2014
Back to Protocols
Resources
- Biological Buffers from Applichem. A good intro, covering all the basics.
General Info
pH = pKa + log[A-]/[HA]
Buffers are usually used within pK ± 1. (source: ISBN 0-19-963142-5 pg 318)
How to chose a buffer for enzyme assays
Information from ISBN 0-19-963142-5 pg 321.
1) Chose the pH range you want. Use a buffer that is within pK± 1 because the buffering capacity will be low outside that range. The range that is used may be varied if it is known that a buffer has only to counter the effects of acid or base but not both. (Is only H+ produced?) The edge of a working range can be more satisfactorily used if the change in pH is towards the pK. 2) Stability of the buffer, whether it interacts with the substrates, cofactors, or metal ions, the temperature coefficients of its pK, the ionic strength at which it is used, its absorbance in the UV region of the spectrum, its cost, and its availability free from contaminants.
Most of the newer zwitterionic buffers do not appreciably bind divalent metal ions, are chemically stable, do not appreciably absorb light at wavelengths longer than 240nm, and can be made up as a concentrated stock solutions.
Many of the buffers which have been in longer use have one or more disadvantages. They are generally cheaper, and if required in large quantities, e.g. for dialysis or column chromatography, may be used provided they have been tested or are known not to affect the enzyme in question. The following drawbacks associated with certain buffers should be borne in mind.
- Phosphate
- Phosphate buffers tend to precipitate Mg2+, Ca2+, Fe3+, and other polyvalent cations.
- Phosphate being an important metabolite is known to inhibit a number of enzymes; e.g. kinases, dehydrogenases, carboxypeptidase, fumarase, urease, aryl sulphatase, adenoside deaminase, phosphoglucomutase, and other enzymes involving phosphate esters.
- On the other hand, it may stabilize enzymes, e.g. phosphoribosepyrophosphate synthase.
- Pyrophosphate
- Pyrophosphate buffers also precipitate polyvalent cations and have a tendency to form complexes. Like phosphate it is also a metabolite and may affect certain enzymes.
- Imidazole
- Imidazole has on occasions been used where phosphate is not acceptable, since both have similar buffering ranges. It is not generally a good alternative, since it is reactive, unstable, and also complexes with divalent metal ions. Mops or Bes might be a better alternative.
- Borate
- Borate has the disadvantage of complexing with vic diols, which included many carbohydrates and the ribose moitey of nucleotides. It would be best avoided when assaying enzymes using NAD(P) or other nucleotides.
- Tris/maleate
- Tris/maleate has been used to cover a wide span of pH from 5.8-8.6, but it does not buffer uniformly between the two pKs, and maleate absorbs in the UV region of the spectrum.
- Barbituric acid
- Barbituric acid buffer used to be extensively used in the range pH 7-9. Apart from being poisonous and inhibiting oxidative phosphorylation, it has the disadvantages o being unstable and of absorbing in the UV region.
- Citric acid
- Citric acid binds some proteins and also complexes many metal ions. It's pKa values are influenced by the ionic strength of the buffer more than most.
- Tris"""
- Tris, although widely used, has the disadvantage common to other primary amine buffers of being reactive and forming Schiff bases with aldehyde and ketones.
- Glycylglycine
- Glycylglycine binds cetain metal ions; e.g. Mn2+ and Cu2+; it may be degraded by peptidases, it is epxensive, and generally has no advantages over the cheaper Tricine.
- Bicarbonate
- Bicarbonate buffer has the disadvantage of having to be used in a closed system to avoid the loss of carbonic acid as CO2.
Resources
Book about buffers
- Helpful book chapter: protein purification handbook (Calbiochem). It is a little mammalian biased, but good.
- Table of Contents: (subset)
- Dissociation Constants of Weak Acids and Bases . . . . . . . . . . . . . . . . . . . . . . 4
- Henderson-Hasselbach Equation: pH and pKa . . . . . . . . . . . . . . . . . . . . . . . . . 5
- Determination of pKa . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 6
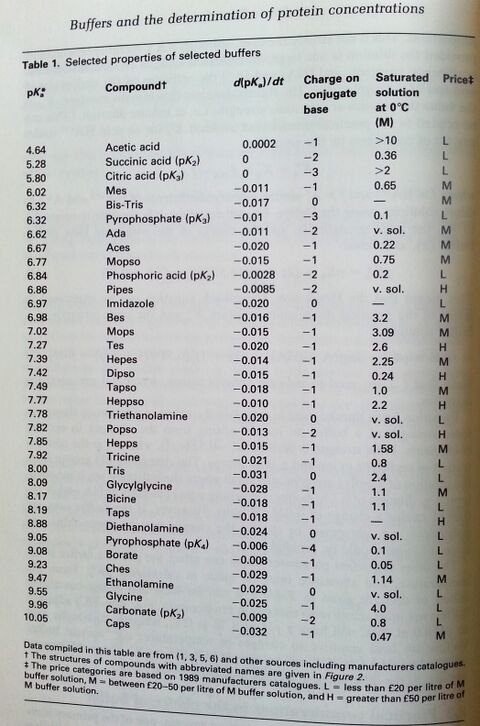
- Values for Commonly Used Biological Buffers. . . . . . . . . . . . . . . . . . . . . . 7
- Buffers, Buffer Capacity, and Range. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 8
- Biological Buffers. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 10
- Buffering in Cells and Tissues . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 10
- Effect of Temperature on pH. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 12
- Effect of Buffers on Factors Other than pH. . . . . . . . . . . . . . . . . . . . . . . . . . . 13
- Use of Water-Miscible Organic Solvents. . . . . . . . . . . . . . . . . . . . . . . . . . . . . 14
- Solubility Equilibrium: Effect of pH on Solubility . . . . . . . . . . . . . . . . . . . . . 14
- pH Measurements: Some Useful Tips . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 15
- Choosing a Buffer. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 16
- Preparation of Some Common Buffers for Use in Biological Systems. . . . . . . 18
- Commonly Used Buffer Media in Biological Research . . . . . . . . . . . . . . . . . . 22
- Isoelectric Point of Selected Proteins. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 24
- Isoelectric Point of Selected Plasma and Serum Proteins. . . . . . . . . . . . . . . . 27
- Approximate pH and Bicarbonate Concentration in Extracellular Fluids . . . 27
- Ionic Composition of Body Fluids. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 28
- Ionization Constants K and pKa for Selected Acids and Bases in Water . . . . . 28
- Physical Properties of Some Commonly Used Acids. . . . . . . . . . . . . . . . . . . . 28
- Some Useful Tips for Calculation of Concentrations and Spectrophotometric Measurements . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 29
- Good exerpts:
- "Effects of Buffers on Factors Other than pH: It is of utmost importance that researchers establish the criteria and determine the suitability of a particular buffer system. Some weak acids and bases may interfere with the reaction system. For example, citrate and phosphate buffers are not recommended for systems that are highly calcium-dependent. Citric acid and its salts are powerful calcium chelators. Phosphates react with calcium producing insoluble calcium phosphate that precipitates out of the system. Phosphate ions in buffers can inhibit the activity of some enzymes, such as carboxypeptidase, fumarease, carboxylase, and phosphoglucomutase. Tris(hydroxy-methyl)aminomethane can chelate copper and also acts as a competitive inhibitor of some enzymes. Other buffers such as ACES, BES, and TES, have a tendency to bind copper. Tris-based buffers are not recommended when studying the metabolic effects of insulin. Buffers such as HEPES and HEPPS are not suitable when a protein assay is performed by using Folin reagent. Buffers with primary amine groups, such as Tris, may interfere with the Bradford dye-binding method of protein assay. Borate buffers are not suitable for gel electrophoresis of protein, they can cause spreading of the zones if polyols are present in the medium"
- Table of Contents: (subset)
- Note Janet hasn't answered the original question! In fact, this pdf only discussed sodium phosphate buffer.
Q&A
Does pH of a buffer depend on the concentration of buffer?
A buffer would be expected to maintain its pH upon dilution, if both [A-] and [HA] are reduced in equivalent proportions. This is not strictly the case, although it is a useful approximation provided the dilution is not large. A discussion of ionic strength follows, informing you that Ka depends on the ionic strength and hence to some degree on dilution. They provide an equation for calculating the effect of dilution or change in ionic strength of a buffer on its pH arising from changes in activity coefficients.
The changes in pH arising from the dilution of a buffer are generally small where the buffering ion is monovalent. Example: dilution of a 0.1M buffer comprising equal amounts of HA and [A-] to 0.05M causes a change of 0.024 pH units. However, if the buffer ions are polyvalent, e.g. phosphate or citrate, the change may be appreciable and large dilutions should be avoided.
(source: ISBN 0-19-963142-5 pg 318)
How does temperature affect the pH of a buffer?
- (source: ISBN 0-19-963142-5 pg 319)
- Amine-containing buffers are most sensitive to changes in temperature. Example: Tris-HCl adjusted to pH=8.0 at 25°C will have a pH of 8.78 at 0°C.
- Carboxylic acid buffers are least sensitive to changes in temperature. Example: acetate buffer adjusted to pH 4.5 at 25°C will have a pH of 4.495 at 0°C.
- These differences are due to the differences in ΔH for ionization of the acids.
- Applichem publication
- The pH value of a Tris solution set to a pH of 7.8 at room temperature is 8.4 at 0°C and 7.4 at 37°C.
How do method developer chose between sodium and potassium phospahte buffers?
- Facts:
- The pKas for sodium and potassium phosphate salts are the same.
- The buffering capacities are the same.
- Potential issues:
- Na or K interferance with an enzyme
- It is possible one will interfere more than the other.
- Solubility
- Sometimes Na precipitates when K wouldn't & vice versa
- Na or K interferance with an enzyme
- Michael Konopka's words: "What assays are you trying to measure? As for the buffering capacity, that really shouldn't matter since the pertinent components for the buffer (mono- and di-basic phosphate) are the same. The issue is if the potassium or sodium ion will form a precipitate which you don't want around if mixing with other solutions (or do want). A classic example is potassium will precipitate SDS while sodium is soluble. That's why in minipreps one adds potassium acetate/acetic acid after using SDS/NaOH to lyse the cells/dissolve lipids & proteins. The proteins, lipids, and chromosomal DNA is then trapped in the precipitate (plasmid DNA still in solution)."
Notes specific to buffers
phosphate buffer
- Phosphates, for example, form insoluble salts with bivalent metals and precipitate. Phosphate buffered salt solution (PBS) is never
autoclaved with Ca2+ or Mg2+ for this reason. Good buffers, such as PIPES, TES, HEPES and CAPS have very low metal-binding constants and are therefore particularly suited to investigate metal-dependent enzymes (Good & Izawa 1972, Blanchard 1984). Applichem