J'aime C. Moehlman's Week 8
From OpenWetWare
Jump to navigationJump to search
Analyzing Protein Structure
- This week we are using the Bioinformatics for Dummies book in order to analyze the protein structure of gp120.
Chapter 2: Retrieving Protein Sequences for the HIV gp120 Envelope Protein
- Searched the protein knowledge base for the gp120 protein.
- Picked envelope glycoprotein 120 with the accession number of A0A897, its entry name is A0A897_9HIV1.
- We retrieved the FASTA sequence for the fragment:NLTDNAKTIIVHLNESVEINCTRPFNNTRTSXRIGPGQVFYRTGDITGSIRRAYCEINGT
KWNKVLXQVTEKLXEH
- We then conducted an advanced search to retrieve a protein sequence.
Chapter 4:Reading a SWISS-PROT entry
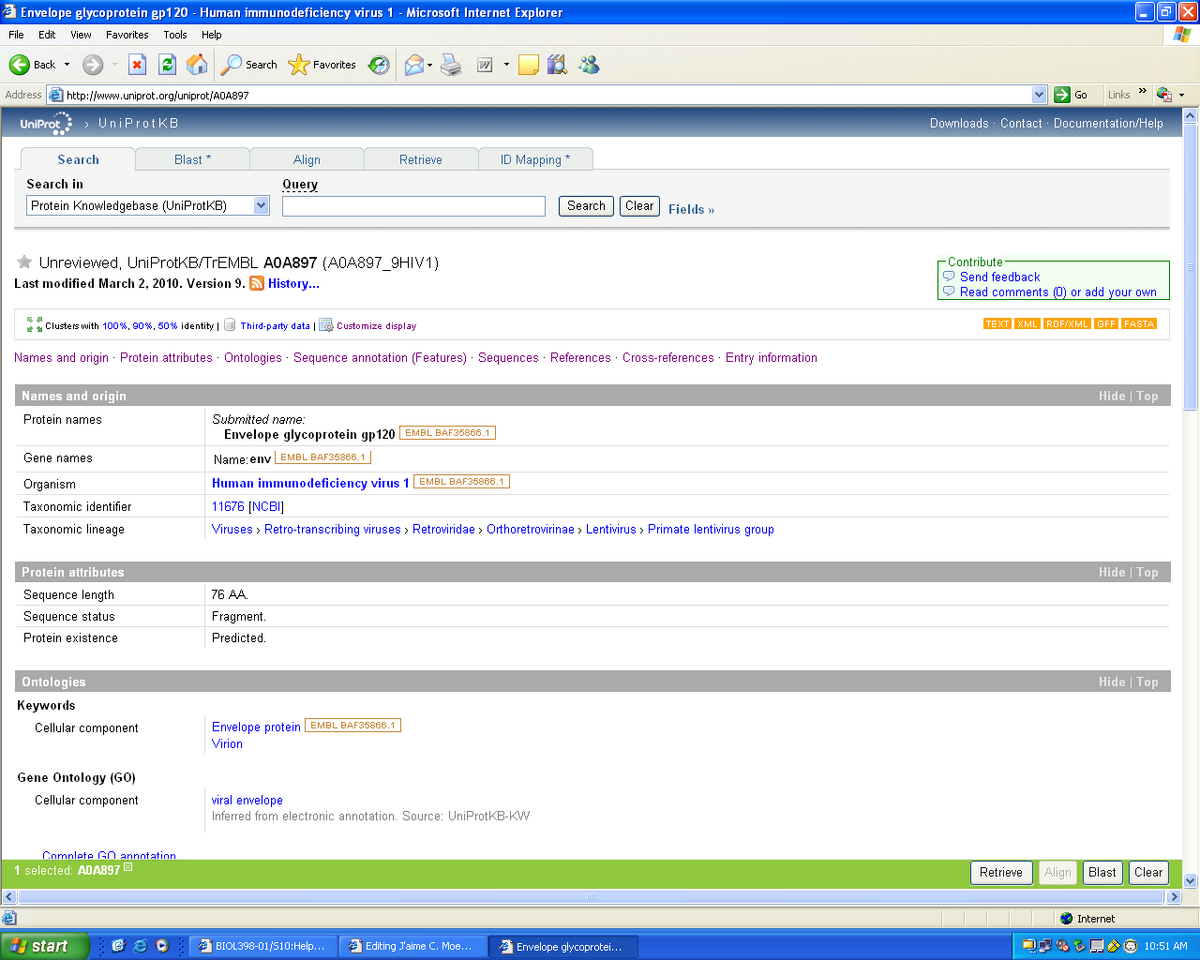
- As seen in the screen shot above we know that the protein name is envelope glycoprotein 120
- The gene name is env
- We looked at the reference section and it shows that there is only one reference which doesn't imply major significance.
- The cross reference page link to entries within other databases:
- Screen shot from EMBL sequence database:
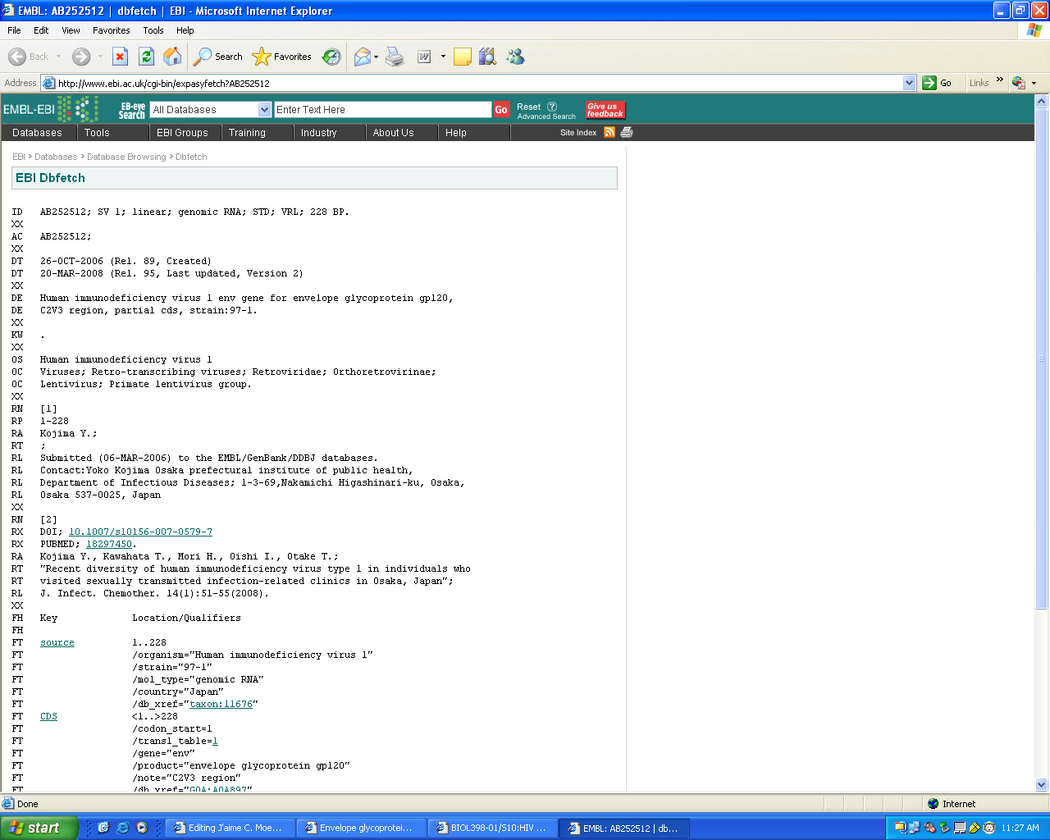
- The keywords for the gp120 protein are; envelope protein and virion.
- In the features section we see non-terminal residues at positions 1 and 76.
Chapter 5:ORFing your DNA sequence
- Picked the DNA sequence from Subject 10 at visit 4 at clone 1.
- SCREENSHOT for ORF:
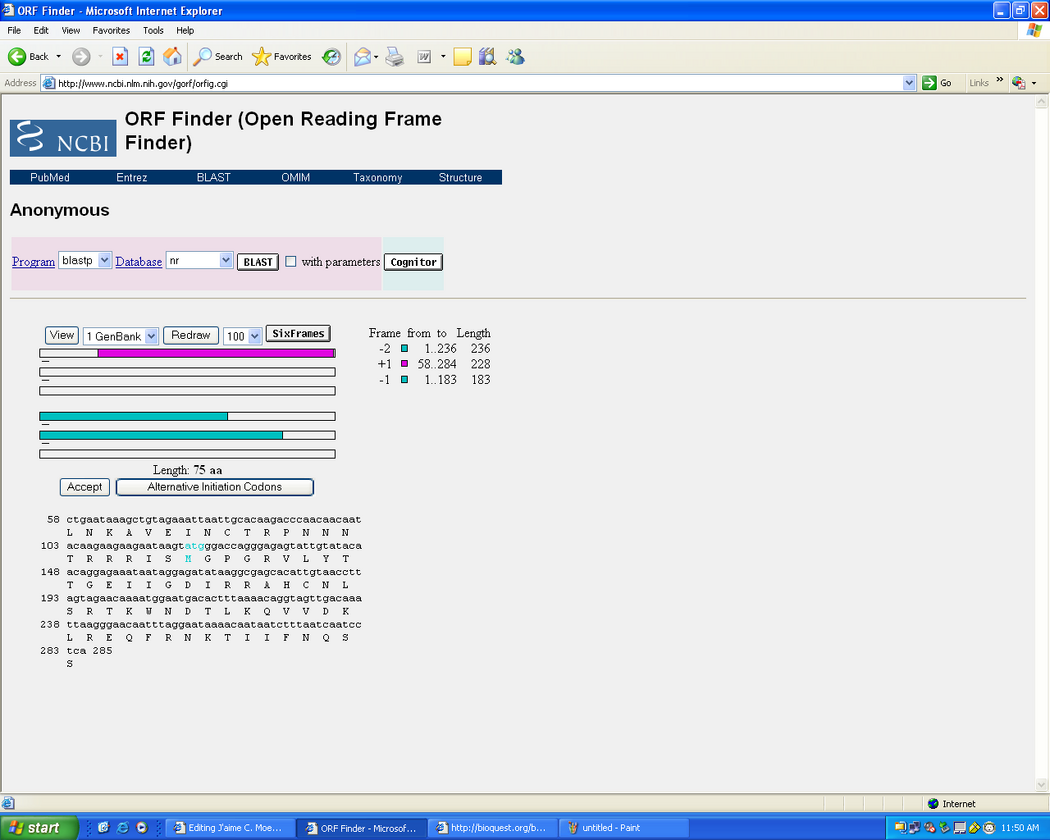
- compared to the SWISS-PROT entry the orf sequence shows very small differences in the sequence.
Chapter 6: Working with a Single Protein Sequence
Predicting the main physico-chemical properties of a protein
- We used the expasy tool page in order to carry out a primary structure analysis.
- We used the accession number A0A987 from Swiss-Prot into the ProtParam.
- ProtParam generated the parameters of the entire gp120 sequence that we selected.
- The results include many things, such as; how many amino acids there are in the sequence, the molecular weight, the overall number of positively charged residues, and the total number of negatively charged residues.
Digesting a protein in a computer
- We pasted the gp120 seguence into the ExPasy website again in order to cut the protein.
Looking for transmembrane segmenting
- We used the accession number A0A987 again and put it into the ExPASy ProtScale site and conducted a full range analysis.
- The image was retrieved in GIF format.
Interpreting ProtScale results
- A piece of paper was used to help us locate the strongest peaks on the graph.
- We determined that there were four important transmembrane regions.
Running TMHMM
- We generated the TMHMM results by using a FASTA sequence of the gp120 protein.
Looking for PROSITE patterns
- Used the accession number A0A897 to determine which proteins we wanted to be scanned and then started the scan.
Finding domains with InterProScan
- We again used the same gp120 protein segment that we have been using throughout the entire assignment.
- We used the our class textbook to interpret the results that were generated by the InterProScan.
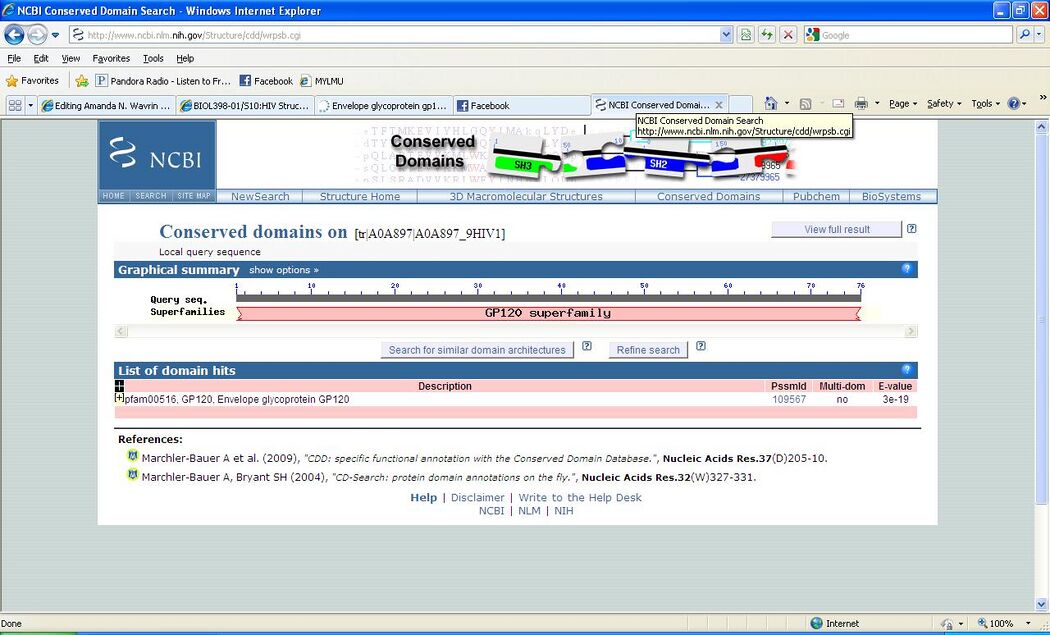
Finding domains with the CD server
- We used the same FASTA gp120 protein sequence.
- This image shows some of our results:
Finding domains with Motif Scan
- We pasted the FASTA sequence into the Input box.
- Selected PROSITE to conduct the search that we wanted.
- With the results, we saw different things: including a matches map, a list of matches, and a matches details section.