ISEEM Progress June 2010
| Home | Project | People | News | For Team | Calendar | Library |
I. Progress made toward completion of our goals
A. Personnel
There have been no personnel changes this quarter.
A rotation student (Aram Avila-Herrara) in the Pollard Lab has been working on iSEEM related projects, including setting up a database of GOS and other CAMERA data locally and mining these data sets for CRISPR diversity (see below). Aram is not paid by the grant.
B. Research
Eisen lab
Protein Classification Pipeline
Tom Sharpton, Dongying Wu, Morgan Langille, and Guillaume Jospin have worked together to cluster all proteins from over 1800 microbial genomes into protein families. To date, they have clustered over 345,000 distinct families and are currently in the process of building probabilistic sequence profiles that describe the evolutionary diversity for each family. These profiles will subsequently be used for a variety of purposes, including metagenomic read annotation. In addition, The family details (members, paths to alignments, size or profile paths) are stored in a MySQL database.
Taxonomic Phylogenetic Marker Identification
We've developed a Shannon Entropy based measurement to estimate how a marker candidate performed on a phylogenetic tree. We call the measurement monophyletic value, and it is scaled from 0 to 100. A monophyletic value of 100 indicates that a marker candidate genes from a specific phylogenetic group form a monophyletic clade on a phylogenetic tree. We've built phylogenetic trees for all the taxonomic specific marker candidates and their top 300 hits outside the correspondent taxonomic groups, and estimate the markers universality, evenness and monophyletic values. We've refined the marker collections as a result.
Table: Phylogenetic Markers for different taxonomic groups Phylogenetic group Genome Number Gene Number Maker Candidates Archaea 62 145415 102 Actinobacteria 63 267783 136 Alphaproteobacteria 94 347287 142 Betaproteobacteria 56 266362 294 Gammaproteobacteria 126 483632 141 Deltaproteobacteria 25 102115 44 Epislonproteobacteria 18 33416 446 Bacteriodes 25 71531 179 Chlamydae 13 13823 561 Chloroflexi 10 33577 140 Cyanobacteria 36 124080 532 Firmicutes 106 312309 80 Spirochaetes 18 38832 72 Thermi 5 14160 727 Thermotogae 9 17037 646
From the taxonomic specific markers, We've identified 209 universal markers that are good markers for at least 5 prokaryotic taxonomic groups (for each group, universality*evenness*monophyletic_value >= 729,000). The markers and their performance for different taxomoic groups are illustrated in the following Figure. The heatmap is based on universality*evenness*monophyletic_value.
Coestimating phylogeny, linkage, and relative abundance in next-generation metagenome data using Bayesian statistics
Aaron Darling developed a new method to directly estimate which metagenome sequence reads come from the same organism, along with the phylogenetic relationship of that organism relative to others in the sample, and also the relative abundance of the organism's DNA in the sample.
The method represents a departure from the standard metagenomic pipeline approach. The standard approach can be summarized as:
- For each read individually:
- Align the read to a most-similar reference family
- Place the read on a phylogenetic tree based on the alignment
- After all reads have been placed:
- Compute OTUs and relative abundance
The new method differs in that it analyzes all reads at once. It can be summarized as:
- Align all reads to their most similar reference families
- Simultaneously estimate:
- linkage groups -- Groupings of reads that come from the same organism
- The phylogeny of linkage groups and reference organisms from the alignment
Each linkage group contains a set of reads that are considered to be related by a phylogenetic star tree topology with very short branch lengths. If the reads all come from the same genome and there is no sequencing error, the true branch lengths should be zero, but I lengthen the branches to accommodate some sequencing error. The star topology within each linkage group connects to the rest of the phylogeny (reference organisms and other linkage groups) from the root of the star.
The estimation of linkage groups and phylogeny is currently computed using Bayesian Markov-chain Monte Carlo techniques. MCMC is a standard statistical technique for taking samples from a probability distribution where direct sampling is difficult, but generating a new sample based on a current sample is easy. Current MCMC updates include all the standard phylogenetic tree topology updates implemented in the BEAST software package, along with a new update that picks a metagenomic sequence read uniformly at random and moves it to another linkage group, also chosen uniformly at random.
In this way, we can sample from the posterior probability distribution of linkage groupings among reads and the phylogeny of linkage groupings and reference organisms. The linkage groupings contain information about the number of reads in the group, which can yield a relative abundance estimate for that linkage group.
By virtue of using Bayesian MCMC, the method assigns a posterior probability that reads are placed together in linkage groups, which gives a measure of statistical confidence in the groupings and the placement of those groupings on the phylogenetic tree.
Some initial tests on the model have been completed using simulated datasets. A dataset was generated based on an amino acid alignment of the rpsB gene from 1727 organisms. The alignment is 220 columns long. 50 organisms were sampled uniformly at random without replacement for use as reference taxa. Another 20 were sampled as metagenome organisms. From those 20 organisms' rpsB gene, 5 sequence reads of 55aa each were simulated. This gives a uniform relative abundance distribution of organisms in the simulated metagenome. Two alignments were saved: an alignment of the 50 reference taxa and the 20 full length metagenome sequences, and the alignment of the 50 reference taxa and the 100 metagenome reads. Standard phylogenetic MCMC was performed on the first alignment, and our new model was applied to the second alignment. The results are shown below.
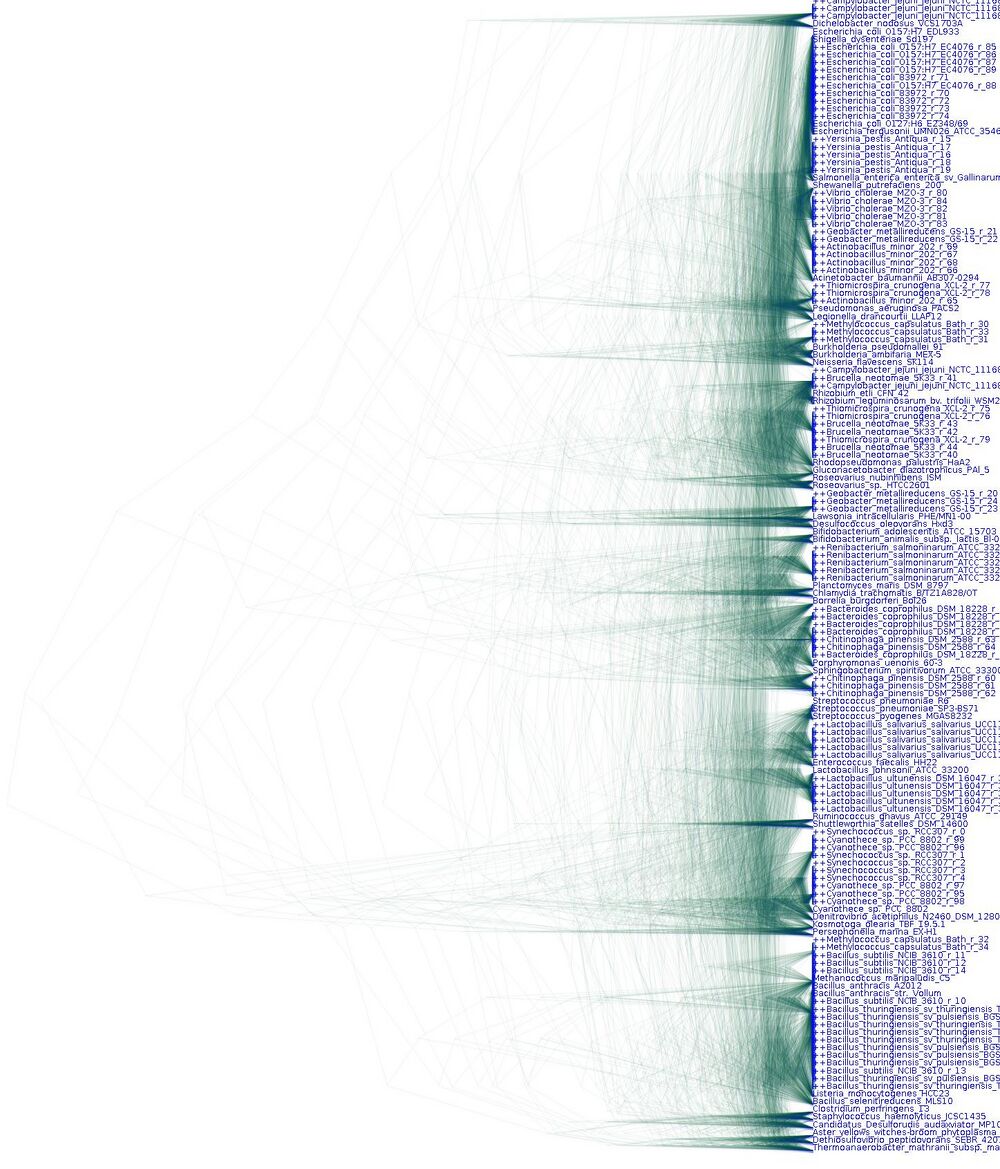
The posterior distribution of trees is shown here. Each tree sampled during the MCMC run is drawn translucent. When many of the sampled trees agree in topology and branch length, they stack on top of each other, creating a darker line. Thus darker lines indicate very confident estimates of tree topology and branch length.
As above, but also including the metagenome reads. Read sequences have names prefixed with ++. It can be seen that reads from the same organism generally cluster together.
Finally, it should be emphasized that these are very early results, and that even though the initial results are extremely encouraging, it is difficult to say how well this method will work in practice.
Characterization of Genes with Unknown Function
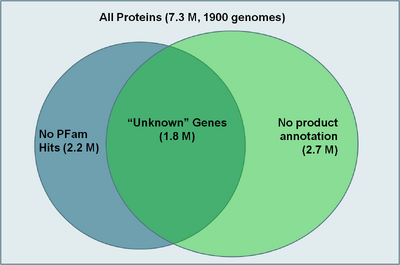
To begin characterization of genes with unknown function they first had to be identified. One method we used was to search for proteins that did not have any PFam hits or only had hits to non-informative PFams such as DUFs (Domains of Unknown Function) and UPFs (Unidentified Protein Families). Using HMMER 3 and all PFAM families we identified 2.2 million proteins that met this criteria (see figure at right). However, some proteins may have a functional annotation, but not have a PFam because a PFam doesn't exist yet for that protein family. Therefore, we searched for genes that did not have a useful product description (e.g. "hypothetical protein", "predicted protein", etc. ) and found 2.7 million proteins that met this criteria. Due to many proteins not being annotated because their genome was still in draft stage, it was decided to take the intersection between these two groups as the list of "unknown genes". This list contains 1.8 million proteins out of the possible 7.3 million proteins for all sequenced genomes.
Using the protein families that we constructed (see "Protein Classification Pipeline" above), we searched for families that contained only (or at least 70%) of genes from our unknown genes list. These protein families with unknown function were then characterized and ranked using several metrics including size, universality, pathogenicity, and aquatic habitat. Based only on the number of members in the unknown families, 10 were found to be very large with greater than 1000 proteins per family. Also, interesting is that 6 families were found to be in all 3 domains of life (Archaea, Bacteria, and Eukaryota), having a universality metric of at least 10% for each family. Considering that pathogens are of great interest there is usually much attention focused on identifying new virulence factors. Our analysis shows that 75 families with with no known function and at least 100 proteins per family were found to contain mostly members from species that are known to be pathogenic. Lastly, 12 families that contain mostly proteins that exist in aquatic species and have at least 50 proteins in each were identified.
The construction and characterization of protein families with no known function has been completed for those sequences coming from completed genomes. Greater information can be obtained by searching for these particular families across many metagenomic datasets of varying ecologies. This will help characterize and hopefully allow for further prediction of what function these families may have.
Green Lab
Using phylogenetic marker genes to measures phylogenetic diversity from metagenomic data
In collaboration with G. Jospin and other collaborators we have applied the phylogenetic marker gene candidates identified by D. Wu to the GOS data set, identifying an additional 839,000 sequences from 116 bacterial and archaeal phylogenetic marker gene families from the metagenomic data.
With the ability to identify hundreds of thousands or millions of sequences from phylogenetic marker gene families in metagenomic data sets, it has become difficult to conduct a single combined analysis of phylogenetic diversity in these data sets. We have explored two approaches to dealing with this issue.
First, we are exploring different approaches for extending existing software to allow measurement of phylogenetic diversity for phylogenies with orders of magnitude more tips than can presently be analyzed. Second, we have conducted analyses where separate analyses of phylogenetic diversity based on multiple phylogenetic marker gene families are combined through averaging and consensus based methods. For example, across the 31 bacterial gene families included in AMPHORA, we found highly consistent patterns of phylogenetic diversity along environmental gradients in the GOS data set, with the same environmental variables explaining most variation in phylogenetic beta diversity across all 31 gene families.
Table. Proportion of total variance in phylogenetic beta diversity (UniFrac distance)
in GOS data set explained by different environmental variables based on analysis
of 31 phylogenetic marker genes included in AMPHORA. Variables explaining at
least 5% of total variation are presented.
Mean SE
Habitat 0.29 0.1
Geographic Location 0.14 0.02
Salinity x Geo. Location 0.07 0.02
Temp x Geo. Location 0.07 0.05
Depth x Geo. Location 0.07 0.01
Salinity 0.05 0.04
Modeling Biodiversity across Environmental Gradients (O'Dwyer)
In our work on the iSEEM project, we have adapted a set of tools from theoretical and statistical physics known as 'field theory', and developed these tools into a new theoretical framework for community ecology. So far, we have explored two distinct applications of these methods. We began with communities structured by body-size (O'Dwyer et al (2009) PNAS), where the size of an organism is taken to be the dominant factor in determining its demographics. This follows an existing body of work in ecology known as metabolic theory, a collection of theoretical derivations meshed with empirical data, telling us that body-size is a key determinant of metabolic rates, from unicellular organisms up to the largest mammals on tree of life. Our work allowed us to combine these ideas from metabolic theory with stochastic population dynamics, and we used this combination to make novel community level predictions.
The second strand of this framework introduces explicitly spatial processes, involving the dispersal or diffusion of organisms across a spatial environment (O'Dwyer & Green (2010) Ecology Letters). The spatial structure of biodiversity is crucial in determining potential shifts in the structure and function of ecological communities in response to environmental change, and investigating this spatial structure has been a major focal point of empirical ecology from its inception. Our work allows us to predict one of the classic patterns of spatial biodiversity, the Species-Area relationship. This relationship characterizes the non-linear increase in diversity with area sampled, and our relatively simple model predicts the shape of this relationship as a function of dispersal capability and speciation rate.
Since the last progress report, we have been working on including a further crucial ingredient: the explicit impact of environmental variability across space. Ultimately we would like to predict the effect of environmental heterogeneity on patterns of taxonomic diversity, and in parallel predict patterns of functional diversity. Our models capture the broad-scale phenomena, but in order to say something more concrete about changes in diversity and function across an environmental gradient, we need to know how this environmental variability feeds into community-level processes.
A classic example of an ecologically-relevant environmental gradient is temperature. For example, there are substantial variations in temperature across both latitude and altitude, with important consequences for biodiversity. This variation illustrates how an environmental gradient can shape both variation in function, with different types of organisms optimized for life in different ranges of temperature, and also patterns of taxonomic and phylogenetic diversity, with an increase in biodiversity with temperature documented in hundreds of studies of the latitudinal and altitudinal diversity gradients. Over the past several months we (O'Dwyer and Green) jointly supervised the thesis research of UO Undergraduate Eric Zaneveld, working with Eric to set up a simple model to capture the impact of a temperature gradient. As a first step we again drew from the body of research on metabolic theory, which shows a strong dependence of metabolic and demographic rates on temperature, and we assumed that species were confined to a very narrow range of temperatures. Building these assumptions into our existing theoretical framework was enough to demonstrate that metabolism can drive patterns in latitudinal diversity similar to those observed empirically. These initial findings are documented in Eric's senior thesis.
We plan subsequently to generalize this framework in a number of directions. First, we would like to include more realistic ranges of temperature tolerance for organisms, and allow adaptation in temperature tolerance over evolutionary time-scales. More generally, we will investigate whether the example of temperature variation can be leveraged to model other environmental gradients, and their impact on correspondingly more general traits---and ultimately we would like to go beyond this to hypothesize and test the impact of particular environmental variables on functional genes, or suites of genes. A model of community assembly which takes into account the impact of environmental variability on taxonomic and functional diversity will be crucial in addressing these questions, and we plan to leverage patterns revealed by metagenomic data to guide the development of these more general theoretical models.
Pollard Lab
PHYLOTU
In collaboration with his co-authors, Tom Sharpton has developed a novel method that quantifies the number of microbial OTUs, or species, from metagenomic data. We have shown that this method, which is dubbed PHYLOTU, can
- accurately, relative to full-length genetic markers, cluster metagenomic reads into OTUs,
- clusters sequences into OTUs similar to traditional, percent identity-based methods, and
- clusters sequences into OTUs in a taxonomically informative mannar.
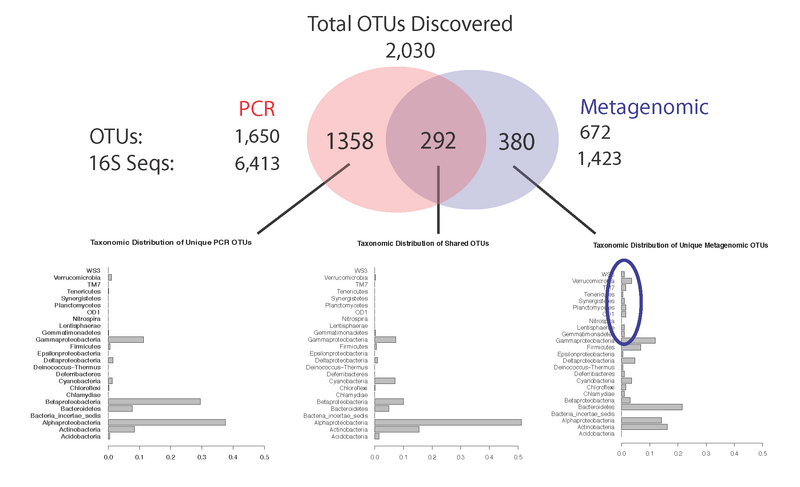
In addition, the entire Global Ocean Survey (GOS) data has been processed by PHYLOTU (metagenomic reads and PCR sequences), which resulted in the discovery of novel microbial taxa at the level of species, genus and family (image below). We have prepared a manuscript describing our results and are preparing to submitt it to PLoS Computational Biology.
The following figure illustrates the results of using PHYLOTU to identify OTUs from all PCR-generated and metagenomic-generated 16S sequences derived from 6 of the GOS samples (the only samples with both PCR and metagenomic 16S sequences). While both types of sequence data identify a common core of OTUs (N=292), both also reveal OTUs unique to each sequence type (N=1358 for PCR-generated sequences, N=380 for metagenomic-generated sequences). We characterized the taxonomy of each set of OTUs via the RDP taxonomy classifier (bar plots below Venn Diagram). We find that using PHYLOTU to analyze metagenomic 16S sequences identifies OTUs from major bacterial lineages that are missed when solely using PCR-based 16S sequences (blue circle).
Distance-Decay and Taxa-Area Relationships
- Distance-decay project
We have completed and submitted the manuscript detailing the piecewise quadratic model of distance-decay. We have extensively revised the manuscript, and improved the model and analyses to better reflect the sampling designs of ecological and metagenomic studies.
- Range estimation project
We have continued developing mathematical proofs to show that our estimators of range shapes are unbiased. These proofs draw on mathematical results from convex geometry and geometric probability.
- Taxonomic diversity estimation project
An ongoing goal has been to develop estimators of taxonomic richness at differing spatial scales, which for instance, will allow us to infer how many microbial OTUs inhabit the North Atlantic Ocean. We have been developing a new estimator of taxonomic richness based on the geometric probability approach developed in the distance decay manuscript. This estimator will complement the other estimators that we have developed, being useful in situations where the other ones are inappropriate.
- Phylogenetic diversity estimation
Biodiversity is quantified not just by taxonomic richness, but also by phylogenetic diversity. We have started working on developing an estimator of phylogenetic diversity at varying spatial scales. This estimator is based on the observation that there may be consistent relationships between phylogenetic diversity and taxonomic diversity. If these relationships exist, then the by quantifying them, we can utilize our taxonomic richness estimators to infer phylogenetic diversity.
Comparison of Phylogenetic Methods for Metagenomics Data
With the help of collaborators, Sam Riesenfeld has finished generating and analyzing the first complete series of simulations for the rpoB protein family. These simulations were designed to test the effects of several variables, including read length, size and diversity of the reference database, number of reads, and phylogenetic inference algorithm, on the accuracy with which simulated metagenomic gene trees recapitulated full-length-sequence gene trees.
Topological error was measured using the normalized Robinson-Foulds metric, which is between 0 and 1. It has been shown that, with very high probability (for natural tree distributions), two random trees with at least 30 leaves have a normalized Robinson-Foulds distance of at least 0.99.
Our main findings include:
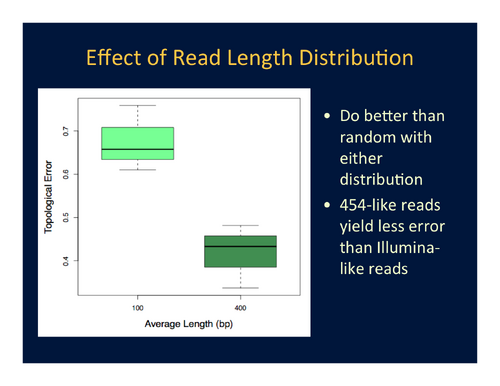
- Topological accuracy is significantly improved by using a 454-like read-length distribution (normal distribution, mean ~400bp, std. dev. ~80bp) rather than an Illumina-like read-length distribution (normal distribution, mean ~100bp, std. dev. ~20bp). The shorter reads do pick up some phylogenetic signal, but it is not strong. See Figure 1.
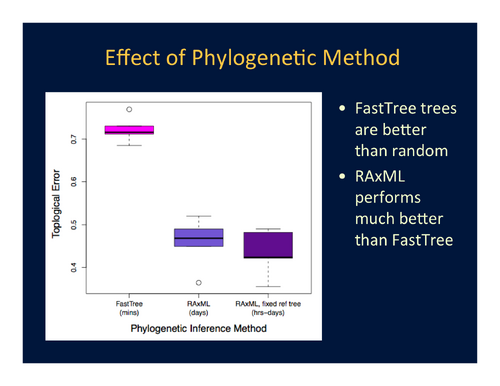
- Topological accuracy is significantly improved by using RAxML (with or without a fixed reference tree), a maximum-likelihood-based phylogenetic inference method, rather than FastTree, a phylogenetic inference method based on Neighbor Joining. However, a RAxML job may take on the order of hours or days, while FastTree finishes in less (often, much less) than half an hour. See Figure 2.
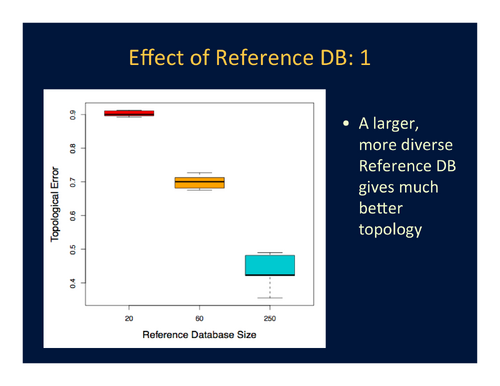
- Topological accuracy is immensely improved by using a reference database that includes almost half of the universe of sequences and is very phylogenetically diverse over reference databases that contain at most 10% of the sequences in the universe, even if the smaller reference databases are relatively phylogenetically diverse. See Figure 3.
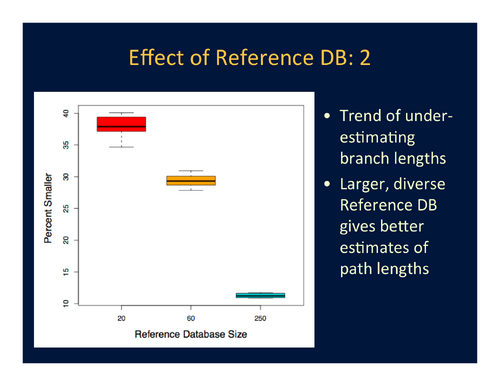
- Branch-length estimation error is widespread and appears to be consistently biased in the same direction for all branches, which means that multiplying all branch lengths by a scaling factor may help address this problem. The problem appears to be reduced when the reference database is large and phylogenetically diverse. See Figure 4.
All of these results were discussed in a platform presentation at the Biology of Genomes Meeting at Cold Spring Harbor Laboratory in May 2010.
Currently, we are working on generating a second series of simulations for both rpoB and 16s rRNA (for which simulations were generated for the PhylOTU analysis) with choices of parameter values that are informed from the results of the first round. We are also adding new protein families to see how similar the results are among the protein families. The first family for which simulations have been generated is nusA, a housekeeping gene, and it appears from these simulations that an additional step of quality control on the alignment should be added. Once simulations for these families are complete, we may add another protein family that does not have a housekeeping function and is hence likely to be dissimilar from phylogenetic marker genes. The results of these simulations will be submitted to a journal for publication. We are also planning a summer submission of an applications note to publish the simulation pipeline itself.
The results of this work have implications for several iSEEM projects, including the PhylOTU project and projects based on comparisons of Phylogenetic Diversity. Generally, the indications are that phylogenetic assessments of metagenomic gene family data should be cautious and attempt to aggregate as much signal as possible.
CRISPRs in Metagenomic Data
Tom Sharpton and a UCSF rotation student (Aram Avila-Herrera) have developed a tool to identify CRISPRs in metagenomic libraries. CRISPRs are repetitive elements in microbial genomes that provide cellular resistance to viral phages cohabiting the ecosystem. CRISPRs have been implicated as important evolutionary and ecological factors, but relatively little is known about them, especially their diversity and natural variation. By screening metagenomic data for their diversity and distribution across ecosystems, we hope to improve the characterization and identify environments where they play especially important roles.
Wu Lab
Project 1
- We performed Ecotype Simulation (ES) analyses of Prochlorococcus sequences from the GOS metagenome using each of the thirty-one housekeeping genes in ComboDB. Demarcations were done based on the parameter solutions for five of those genes. Each analysis demarcated multiple putative ecotypes within Prochlorococcus marinus, three of which seem to correspond with subclades of that taxon previously designated as ecotypes by other researchers, based on associations with different temperatures. However, we have not yet been able to detect any definite associations between the ecotypes we have demarcated, and any habitat parameters in the GOS metadata. The number of ecotypes demarcated still seems to vary greatly depending on the gene that we are analyzing (e.g. infC, pgk and rpoB each predict thirteen to fourteen ecotypes, while dnaG and pyrG each predict over seventy ecotypes. We have been experimenting with different metrics for approximating the rate of neutral sequence evolution in these genes to test the hypothesis that more quickly evolving genes will return higher rates of ecotype formation and especially periodic selection, and therefore return a higher estimate for the total number of ecotypes.
- Ecotype Simulation is also being used to analyze Pelagibacter 16s sequences analyzed by Acinas et. al. (Acinas et. al. 2004). These sequences, obtained by PCR rather than metagenomic seqeuncing, are more conducive to use with the Ecotype Simulation algorithm. We can use representives of predicted ecotypes from this analysis to serve as additional reference sequences for continuing our ES analysis of the Pelagibacter metagenomic data. We are also using them to test different methods of binning sequences to generate the Clade Sequence Diversity Curve, that are potentially less restrictive than the binning algorithm packaged with ES. We also intend to use the results of ES analysis of this and other, larger 16s data sets to compare ES-demarcated ecotypes to OTUs based on universal molecular cut-offs.
C. Communications, Collaboration, Outreach and Education
Publications supported by this grant
- T. Sharpton, S. Riesenfeld, S. Kembel, J. Ladau, J. O'Dwyer, J. Green, J. Eisen, K. Pollard. “PHYLOTU: A high-throughput procedure that identifies Operational Taxonomic Units from metagenomic data.” (In prep; near submission to PLoS Computational Biology)
- J. Ladau, J. Green, K. Pollard. “Beta Diversity Follows a Universal Model.” (Submitted to Theoretical Ecology)
- S.W. Kembel, J.A. Eisen, K.S. Pollard, J.L. Green. Community phylogenetics reveals novel insights into microbial diversity and community assembly. (Submitted to PLoS Biology)
- S.W. Kembel, P.D. Cowan, M.R. Helmus, W.K. Cornwell, H. Morlon, D.D. Ackerly, S.P. Blomberg, and C.O. Webb. 2010. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26:1463-1464.
- Hartman AL, Riddle S, McPhillips T, Ludaescher B, Eisen JA. WATERS: a Workflow for the Alignment, Taxonomy, and Ecology of Ribosomal Sequences. BMC Bioinformatics. 2010 Jun 12;11(1):317.
- Langille MG, Eisen JA. BioTorrents: a file sharing service for scientific data. PLoS One. 2010 Apr 14;5(4):e10071.
Talks
- Jessica Green. "Biodiversity Theory and Metagenomics-Based Biogeography". Stanford Symposium on Evolution and Genomics. April 2010.
- Katie Pollard. "The iSEEM Project: Phylogenetic Approaches to Microbial Metagenomics." Cold Spring Harbor, Biology of Genomes Meeting, May 11-15. Katie was invited to chair the session on non-human genomics at this meeting.
- Sam Riesenfeld. "Building phylogenies with metagenomic sequence reads." Biology of Genomes Meeting, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, May 2010.
- Tom Sharpton. “Charactering Microbial Diversity from Metagenomic Data” Research In Progress Symposium, Gladstone Institute of Cardiovascular Disease, June 2010.
- Tom Sharpton et. al. “Resolving the Hidden Biosphere: A high-throughput procedure that identifies Operational Taxonomic Units from metagenomic data”. Evolution Annual Meeting, June 2010.
- S.W. Kembel et al. "Picante: R tools for integrating phylogenies and ecology". iEvoBio Meeting, June 2010.
Posters
- Josh Ladau, Sam Riesenfeld, Tom Sharpton, Katie Pollard. “Characterizing Microbial Diversity: Who is out there and what are they doing?” J. David Gladstone Institute Annual Retreat, April 2010.
News
- iSEEM Ecology Letters 2010 paper by O'Dwyer and Green was highlighted in the Nature Journal club and won the Ecological Society of America Theory Paper of the Year award.
II. Group meetings
Weekly meetings
- Notes 4_7_10
- Notes 4_14_10
- Notes 4_21_10
- Notes 4_27_10
- Notes 5_5_10
- Notes 5_19_10
- Notes 6_2_10
- Notes 6_9_10
- Notes 6_17_10
Other meetings
- Eugene, OR: June 24-25, 2010. Schedule.
III. Any unexpected challenges that imperil successful completion of the Outcome
CAMERA interactions
Still nothing major to report. We had one phone call with CAMERA over the last few months that related to building a Kepler workflow for the STAP software. They have made some progress in this but not a lot.