IGEM:IMPERIAL/2009/M1
Module 1 - Genetic Circuit
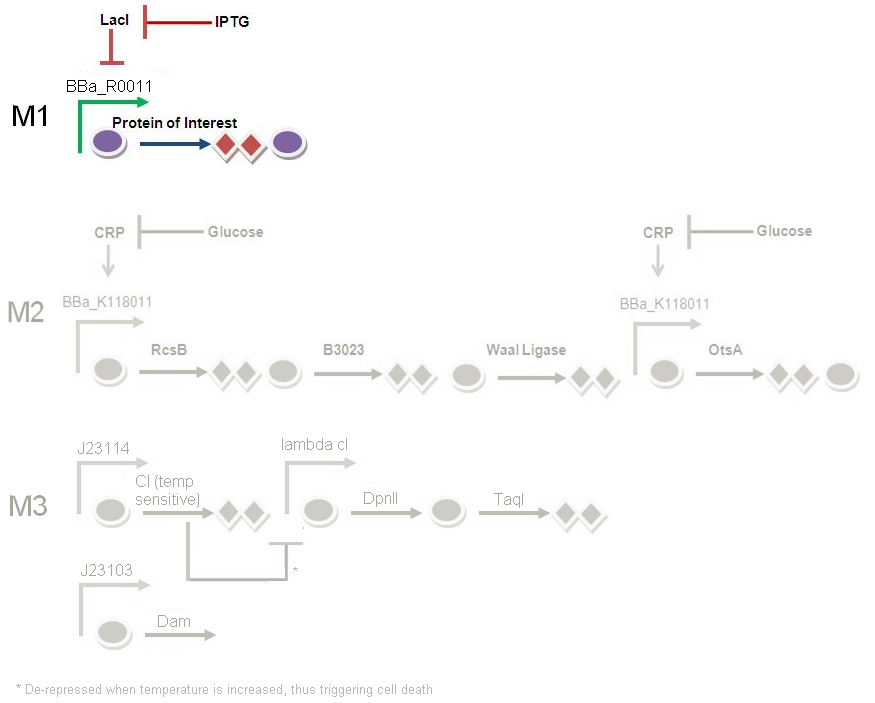
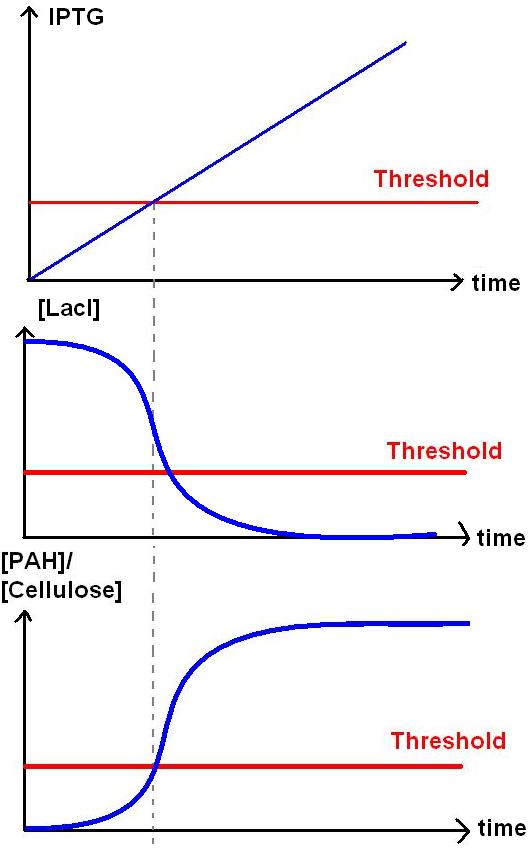
In Module 1 (M1), LacI is constitutively produced. LacI represses the LacI repressible promoter which controls the production of PAH/cellulase.
In order to start off M1, we pipette in IPTG. This represses LacI production, which was originally repressing BBa_R0011. BBa_R0011 is the promoter responsible to kick start the production of PAH / cellulase. As the repression on the promoter is now lifted, the genes under the promoter can now be transcribed to produce the PAH/cellulase.
In conclusion, the addition of IPTG represses the productio of functional LacI, and this will subsequently start protein production.
%note that since LacI is being produced constitutively, when the added IPTG is used up, concentrations of LacI will increase again and enzyme production will stop. We can solve this problem by pipetting in IPTG at regular time intervals. This needs further research.
Module 1 - Protein production
- People: Charles and Tianyi
DNA constructs
This module is aimed at synthesising the protein of interest that will be subsequently encapsulated. We are focusing on two main proteins for this module. The module however can be easily adapted to other proteins.
Phenylalanine Hydroxylase (PAH)
Mutation in this enzyme has been associated with occurence of Phenylketonuria. Because PAH cannot break-down phenylalanine (an amino acid that cannot be synthesised by the body, it has to come from the diet) into tyrosine, phenylketonuriacs accumulate phenylalanine in the blood and in the brain which in term causes mental retardation if the diet is not tightly controlled. More information on PKU here, here and here
--Kun Xue 09:39, 11 August 2009 (EDT)Whats at each 'here'?
- Nuri Purswani 06:14, 12 August 2009 (EDT): Do we need to show the entire DNA sequence? maybe a link to it in a new page will be better
DNA sequence
ATGTCCACTGCGGTCCTGGAAAACCCAGGCTTGGGCAGGAAACTCTCTGACTTTGGACAGGAAACAAGCTATATTGAAGACAACTGCAATCAAAATGGTGCCATATCAC
TGATCTTCTCACTCAAAGAAGAAGTTGGTGCATTGGCCAAAGTATTGCGCTTATTTGAGGAGAATGATGTAAACCTGACCCACATTGAATCTAGACCTTCTCGTTTAAA
GAAAGATGAGTATGAATTTTTCACCCATTTGGATAAACGTAGCCTGCCTGCTCTGACAAACATCATCAAGATCTTGAGGCATGACATTGGTGCCACTGTCCATGAGCTT
TCACGAGATAAGAAGAAAGACACAGTGCCCTGGTTCCCAAGAACCATTCAAGAGCTGGACAGATTTGCCAATCAGATTCTCAGCTATGGAGCGGAACTGGATGCTGACC
ACCCTGGTTTTAAAGATCCTGTGTACCGTGCAAGACGGAAGCAGTTTGCTGACATTGCCTACAACTACCGCCATGGGCAGCCCATCCCTCGAGTGGAATACATGGAGGA
AGAAAAGAAAACATGGGGCACAGTGTTCAAGACTCTGAAGTCCTTGTATAAAACCCATGCTTGCTATGAGTACAATCACATTTTTCCACTTCTTGAAAAGTACTGTGGC
TTCCATGAAGATAACATTCCCCAGCTGGAAGACGTTTCTCAATTCCTGCAGACTTGCACTGGTTTCCGCCTCCGACCTGTGGCTGGCCTGCTTTCCTCTCGGGATTTCT
TGGGTGGCCTGGCCTTCCGAGTCTTCCACTGCACACAGTACATCAGACATGGATCCAAGCCCATGTATACCCCCGAACCTGACATCTGCCATGAGCTGTTGGGACATGT
GCCCTTGTTTTCAGATCGCAGCTTTGCCCAGTTTTCCCAGGAAATTGGCCTTGCCTCTCTGGGTGCACCTGATGAATACATTGAAAAGCTCGCCACAATTTACTGGTTT
ACTGTGGAGTTTGGGCTCTGCAAACAAGGAGACTCCATAAAGGCATATGGTGCTGGGCTCCTGTCATCCTTTGGTGAATTACAGTACTGCTTATCAGAGAAGCCAAAGC
TTCTCCCCCTGGAGCTGGAGAAGACAGCCATCCAAAATTACACTGTCACGGAGTTCCAGCCCCTGTATTACGTGGCAGAGAGTTTTAATGATGCCAAGGAGAAAGTAAG
GAACTTTGCTGCCACAATACCTCGGCCCTTCTCAGTTCGCTACGACCCATACACCCAAAGGATTGAGGTCTTGGACAATACCCAGCAGCTTAAGATTTTGGCTGATTCC
ATTAACAGTGAAATTGGAATCCTTTGCAGTGCCCTCCAGAAAATAAAGTAA
Cellulase
We propose to use a cellulase enzyme able to catalyse all three enzymatic reactions (crystal to cellulose to cellobiose/cellotetrose and to glucose) involved in the degradation of cellulose crystals into glucose. The particular enzyme we will be using is from Clostridium thermocellum Gene/Protein information. Another advantage of using Clostridium's cellulase is that it is protease resistant, therefore will be protease resistant in the target environment of the small intestine. More information can be found here.
DNA sequence:
ATGAAAAAAATTGTTTCTTTGGTTTGTGTGCTTGTGATGCTGGTAAGCATCTTAGGCTCGTTTTCAGTCGTAGCGGCATCACCGGTAAAAGGC
TTTCAGGTATCGGGAACAAAGCTTTTGGATGCAAGCGGAAACGAGCTTGTAATGAGGGGCATGCGTGATATTTCAGCAATAGATTTGGTTAAA
GAAATAAAAATCGGATGGAATTTGGGAAATACTTTGGATGCTCCTACAGAGACTGCCTGGGGAAATCCAAGGACAACCAAGGCAATGATAGAA
AAGGTAAGGGAAATGGGCTTTAATGCCGTCAGAGTGCCTGTTACCTGGGATACGCACATCGGACCTGCTCCGGACTATAAAATTGACGAAGCA
TGGCTGAACAGAGTTGAGGAAGTGGTAAACTATGTTCTTGACTGCGGTATGTACGCGATCATAAATGTTCACCATGACAATACATGGATTATA
CCTACATATGCCAATGAGCAAAGGAGTAAAGAAAAACTTGTAAAAGTTTGGGAACAAATAGCAACCCGTTTTAAAGATTATGACGACCATTTG
TTGTTTGAGACAATGAACGAACCGAGAGAAGTAGGTTCACCTATGGAATGGATGGGCGGAACGTATGAAAACCGAGATGTGATAAACAGATTT
AATTTGGCGGTTGTTAATACCATCAGAGCAAGCGGCGGAAATAACGATAAAAGATTCATACTGGTTCCGACCAATGCGGCAACCGGCCTGGAT
GTTGCATTAAACGACCTTGTCATTCCGAACAATGACAGCAGAGTCATAGTATCCATACATGCTTATTCACCGTATTTCTTTGCTATGGATGTC
AACGGAACTTCATATTGGGGAAGTGACTATGACAAGGCTTCTCTTACAAGTGAACTTGATGCTATTTACAACAGATTTGTGAAAAACGGAAGG
GCTGTAATTATCGGAGAATTCGGAACCATTGACAAGAACAACCTGTCTTCAAGGGTGGCTCATGCCGAGCACTATGCAAGAGAAGCAGTTTCA
AGAGGAATTGCTGTTTTCTGGTGGGATAACGGCTATTACAATCCGGGTGATGCAGAGACTTATGCATTGCTGAACAGAAAAACTCTCTCATGG
TATTATCCTGAAATTGTCCAGGCTCTTATGAGAGGTGCCGGCGTTGAACCTTTAGTTTCACCGACTCCTACACCTACATTAATGCCGACCCCC
TCGCCCACGGTGACAGCAAATATTTTGTACGGTGACGTAAACGGGGACGGAAAAATAAATTCTACAGACTGTACAATGCTAAAGAGATATATT
TTGCGTGGCATAGAAGAATTCCCAAGTCCTAGCGGAATTATAGCCGCTGACGTAAATGCGGATCTGAAAATCAATTCCACCGACTTGGTATTG
ATGAAAAAATATCTACTGCGCTCAATAGACAAATTTCCTGCGGAGGATTCTCAAACACCTGATGAAGACAATCCGGGCATTTTGTATAACGGA
AGATTCGATTTTTCAGATCCGAACGGTCCGAAATGCGCCTGGTCCGGCAGCAATGTTGAGCTGAATTTTTACGGCACGGAAGCAAGTGTGACT
ATCAAATCCGGCGGTGAGAACTGGTTCCAGGCTATTGTAGACGGCAATCCTCTTCCTCCTTTTTCGGTTAACGCTACTACCTCTACCGTAAAG
CTTGTAAGCGGTCTTGCAGAAGGAGCTCATCATCTTGTATTGTGGAAGAGGACAGAGGCATCCTTGGGAGAAGTTCAGTTCCTTGGGTTTGAT
TTTGGTTCAGGAAAGCTTCTTGCCGCACCGAAGCCTTTGGAAAGAAAGATTGAGTTTATCGGAGACTCCATCACATGTGCATACGGAAATGAA
GGAACAAGCAAGGAGCAGTCTTTTACACCGAAAAATGAAAACAGCTATATGTCTTATGCGGCAATTACAGCCCGTAATTTGAATGCAAGTGCA
AATATGATTGCGTGGTCCGGAATCGGACTTACCATGAACTACGGCGGAGCCCCCGGACCTCTTATAATGGACCGTTATCCTTATACCCTTCCT
TACAGCGGAGTCAGATGGGATTTTAGCAAATATGTGCCTCAGGTTGTTGTAATCAATCTTGGTACCAATGATTTTTCTACATCATTTGCAGAT
AAAACAAAGTTTGTAACGGCATATAAAAACCTTATAAGTGAAGTTCGCAGGAACTATCCGGATGCCCATATATTCTGCTGTGTCGGTCCGATG
CTTTGGGGAACGGGCCTGGATTTGTGCCGCAGTTATGTTACGGAAGTTGTAAATGATTGTAACAGAAGCGGGGATTTAAAGGTGTATTTTGTT
GAGTTTCCGCAGCAGGACGGAAGCACCGGATACGGAGAAGACTGGCATCCAAGTATTGCCACCCACCAGCTGATGGCTGAGCGGCTTACTGCG
GAAATAAAAAACAAGCTTGGATGGTAA
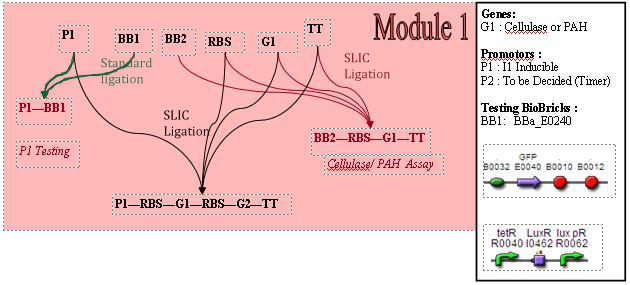
Cloning strategy (assembly)
Testing constructs are in red, and the genetic circuit for M1 is in black.
Testing Constructs
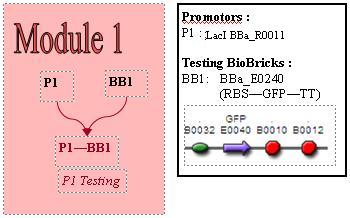
Promoter Testing
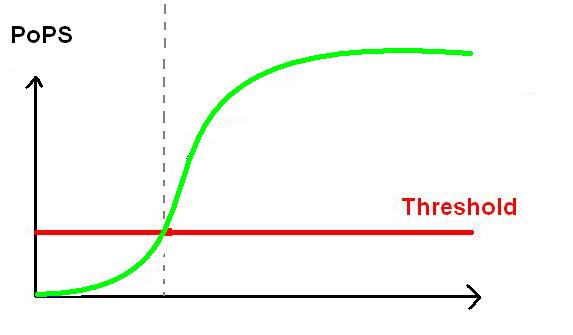
The biobrick used, BBa_E0240 allows us to characterise the promoter. To do so, we attach this biobrick to the end of the promoter we are testing ie, the LacI promoter, BBa_R0011. BBa_E0240 will output GFP. We can indirectly measure the activity of promoters via observing the rates of GFP synthesis. GFP synthesis rate can be estimated by looking at the change in arbiturary fluorescence units per absorbance. Therefore, we'll need a graph of fluorescence vs time. The gradient of the slope of this graph would give rate of GFP synthesis. Subsequently, we can estimate promoter activity, using the equation:
This equation can be further simplified by making many assumptions.
Promoter activity is measured in terms of PoPS. Relative promoter activity will be in relative promoter units (RPU). RPU is measured by using the promoter activity of BBa_J23101 as the standard reference.
(Refer to Jason Kelly's paper)
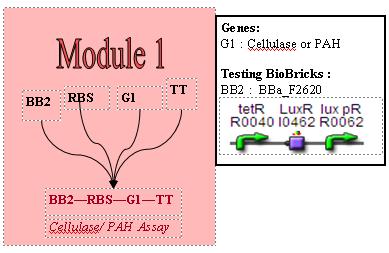
Gene Testing
| INPUT | TESTING CONSTRUCT | OUTPUT |
| HSL (mM) | F2620-RBS-Cellulase Gene-TT (-Assay) | Fluorescence (RFU) |
BB2, BBa_F2620 is a well characterised promoter sequence, hence we will know the exact PoPS output for a certain amount of HSL input. By adding BBa_F2620 in front of our gene of interest, we know the exactly what our input PoPS is. We can then measure our PAH / Cellulase output per unit time using the assays. This will give us our PoPS output. In that sense, BBa_F2620 allows us to measure the behaviour of the genes coding for PAH or cellulase, depending on the input concentration of HSL, and will allow us to characterise our own biobricks
Assays & Protocols
IPTG effect on growth at 28 degrees
Output data
Promoter
- Graph of fluorescence against time for different IPTG input.
- Using the formula in the Jason Kelly paper, we can relate fluorescence to GFP synthesis rate per cell, and then to PoPS
- Nuri Purswani 09:15, 12 August 2009 (EDT): See genetic circuit: inputs Iptg and LacI
PAH
PAH activity: 1 unit = 1 mmol.min-1 @ pH=7.2; 25°C
- Calibration curve of [PAH] vs colour intensity.
Cellulase
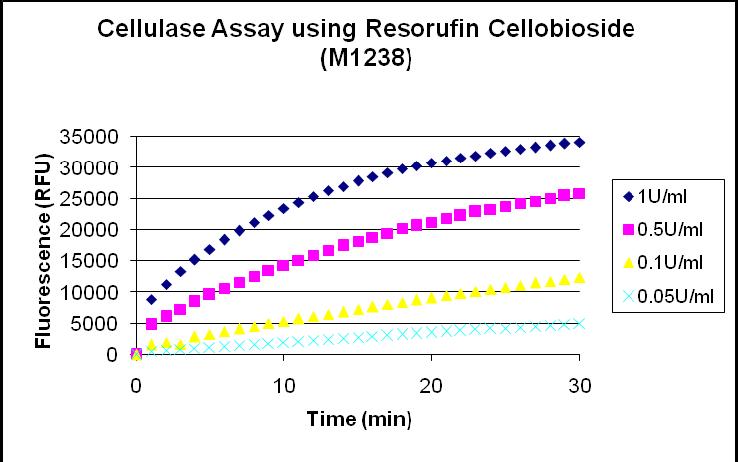
- The assay output will provide us a measure of Relative fluorescence unit (RFU) over time.
- We can directly relate RFU to enzymatic activity(U/ml) using a standard curve (See plot below)
1995 - Cellulase activity: 1 unit = 1 μmol.min-1 @ pH=5.0; 37°C
- The slope of the Fluorescence vs Time curve should be the rate of production of fluorescence. Assuming that only the cellulase enzyme can catalyse the above reaction, the slope should represent the activity of the enzyme
- Nuri Purswani 09:19, 12 August 2009 (EDT): I think that for the modelling in this part, we should work from our assays and testing constructs.
Possible questions we may want to answer: 1. Promoter characterization as a function of LacI, IPTG. 2. Michaelis-Menten kinetics: ODE models of chemical reactions. We want to relate initial concentrations of substrate in our testing constructs to product and characterize enzymatic activity (of PAH and cellulase). To do this we will first go through the 2nd practical of the computer modelling synth bio course. We will then try to break down our system in a similar manner, noting the key questions we would like to ask.
Input from wet-lab
1) Calibration curve to determine [PAH] and [cellulase].
2) When testing the promoter using BBa_E0240, obtain a graph of fluorescence vs time
Modelling
1) Data analysis : Derive rate of production of cellulase / PAH
2) Modelling:
- Cell growth rates
- Determine rate of production of PAH / Cellulase
-LacI is produced constitutively, this repressed M1. -When there is IPTG input, it represses LacI below threshold. M1 will kick start. -When there's above threshold IPTG, there would be production of PAH / Cellulose.
- To characterise any possible biobricks.
- To characterise promoter, plot GFP vs [IPTG] graphs, and GFP vs time graphs etc
- To characterise the gene for either PAH or cellulase, plot graphs of IPTG and LacI (produced constitutively) against assay output (which is either fluorsecence, or colour). Also, perhaps generate a 3D graph of IPTG against assay output and time.
- Relating PoPs output to colour (PAH) or fluorescence (Cellulose)
- PAH / cellulase enzyme kinetics [1]
- Normalisation of data
- Fitting of wetlab data (ie activity vs time) to model
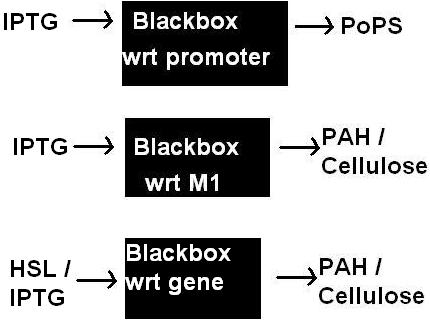
- Transfer functions for
- IPTG --> blackbox --> PoPS
- HSL --> blackbox --> PoPS (testing construct)
- IPTG --> blackbox of M1 --> PAH or cellulase
- HSL --> blackbox of M1 --> PAH or cellulase (testing construct)
- use Drew Endy standardisation for PoPS output, so as to relate it to GFP
3) Ultimate aim: Determine a model that treats M1 as a blackbox. ie. IPTG --M1--> PAH / Cellulase
Shopping list
Cellulase Fluorescence 2005 (preferred)
| Assay kit | Where to buy | Price |
| E33953 EnzChek® Cellulase Substrate *blue fluorescent, 339/452 | invitrogen | £131.00 / 186 assays |
Cellulase Fluorescence 2009
| Assay kit | Where to buy | Price |
| MarkerGene Fluorescent Cellulase Assay Kit | biokit or markergene | £105.49 or £138.23 / kit |
Cellulase Fluorescence 1995
| Reagent | Where to buy | Notes |
| Sodium Acetate Buffer, pH 5.0 | Sigma | 250g, £7.90 |
| Sigmacell Solution (Sigmacell) | sigma | 300g, £30.90 |
| Glucose (HK) Determination Vial (16-10) | sigma@ | 20 assays, £38.80 |
@ - Can be the same kit as M2
Phenylalanine hydroxylase
| Reagent | Where to buy | Notes |
| Trizma Base | sigma | 25g, £10.10 |
| L-Phenylalanine | sigma | 100g, £34.00 |
| Catalase | sigma | 50mg, £43.00 |
| DL-Dithiothreitol | sigma | 1g, £19.40 |
| DL-6-Methyl-5,6,7,8-Tetrahydropterine Dihydrochloride | sigma | 50mg, £69.50 |
| Trichloroacetic Acid, 6.1 N Solution | sigma | 100ml, £31.40 |
| Nitric Acid | sigma | - |
| Sodium Nitrite | sigma | 25g,£59.50 |
| Nitrosonaphthol | sigma | 25g, £27.10 |
| Sodium Hydroxide Solution | sigma | - |
| L-Tyrosine Free Base | sigma | 50g, £13.80 |