Constructs Overview
Here are several tables listing the bricks which we are cloning, constructing and using, with their relevant details.
A new page is currently being developed!
Cloning Table
Status of biobricks cloned and their applications:
I
ID
Part
Description
Status
Ligation Target
Use in Constructs
02
BBa_I13522 ptet-GFP
XL1 , Maxi Mini BL21 ---
Basic construct for chassis characterization
11
BBa_E7104 pT7 GFP
XL1 , Maxi Midi Mini BL21 ---
CBD test construct (pT7)
30
---
pT7 (strong GFP)
XL1 , Maxi Mini BL21 ---
T7 test-construct
I 18
BBa_T9002 ptet-luxR-plux-GFP
XL1 , Maxi Midi BL21 ---
Basic construct for chassis characterization
05
BBa_R0040 ptet
XL1 , Midi Mini DsRedEx
CBD final (ptet), ID AHL sender
BBa_I719???
pT7
Ready DsRedEx
CBD final construct (pT7)
07
BBa_F2620 ptet-luxR-plux
XL1 , Midi Mini DsRedEx
ID final construct
21
BBa_J23039 ptet-luxI
XL1 , Midi ---
ID sender construct
24
BBa_R0062 plux
XL1 , Midi DsRedEx
ID construct 2
25
BBa_J37032 plux-GFP
XL1 , Maxi Midi ---
ID construct 2
26
BBa_S03119 ptet-luxR
XL1 , Midi ---
---
27
pSB1A3 Empty pSB1A3
Problematic ---
---
13
BBa_B0015 Double terminator
XL1 , Maxi Midi Mini ---
General part construction
17
BBa_I13507 RFP
XL1 , Midi ---
Not being used anymore
19
BBa_B0034 strong RBS
XL1 ---
General part construction
23
BBa_I13504 GFP
XL1 , Maxi Midi pT7
General part construction
BBa_I719???
DsRed Express
Ordered
ptet, plux, pT7
Reporter system for ID, Hrp
BBa_I719???
HrpR-HrpS
Ordered
???
Hrp regulators
BBa_I719???
pHrpL
Ordered
???
Hrp promoter
BBa_I719???
HrpV
Ordered
???
Hrp inhibitor system
Ligation Table
Status of parts ligated and their applications:
I
ID
Part
Description
Status
Parents
Application
BBa_I719???
pT7 & GFP
Planned
pT7 + BBa_I13504
CBD test construct (pT7)
BBa_I719???
DsRedEx & pSB1A3
Paused DsRedEx + BBa_P1010
Biobrick Submission
BBa_I719???
pT7 & DsRedEx
Planned
pT7 + DsRedEx
CBD final construct (pT7)
BBa_I719???
ptet & DsRedEx
Planned
BBa_R0040 + DsRedExCBD final construct (ptet)
BBa_I719???
ptet-luxR-plux-DsRedEx
Planned
BBa_F2620 + DsRedExID detector final construct 1
BBa_I719???
plux-DsRedEx
Planned
BBa_R0062 + DsRedExID detector final construct 2
Legend: I (Information), [Status: XL1 - cloned in XL1-Blue; Maxi Midi Mini BL21 - cloned in BL21(DE3); status - stocks running low]
Parts from the Registry
Physical information of biobricks:
A
W
G
S
ID
Name
Description
Plasmid
Resist
Copy
Size(B)
Size(T)
Location
Conc.
A
W G
02
BBa_I13522 GFP under tetracyclin promoter
pSB1A2 AmpR
High
937bp
3016bp
Well 15H, Plate 2
500
A
W
G
05
BBa_R0040 tetracyclin promoter
pSB1A2 AmpR
High
54bp
2133bp
Well 7O, Plate 1
60
A
W
G
07
BBa_F2620 lux promoter, with luxR
pSB1A2 AmpR
High
1061bp
3140bp
Well 6B, Plate 1
80
A
X G 11
BBa_E7104 T7 GFP
pSB1A2 AmpR
High
826bp
2905bp
Well 13F, Plate 2
100
A
W
G
13
BBa_B0015 Double terminator
pSB1AK3 A+KR
High
129bp
3316bp
Well 1B, Plate 2
100/70
A
G
17
BBa_I13507 RFP
pSB1A2 AmpR
High
861bp
2940bp
Well 16N, Plate 1
80
A
W G
18
BBa_T9002 GFP under lux promoter with LuxR
pSB1A3 AmpR
High
1945bp
4102bp
Well 19B, Plate 2
460
A
W
19
BBa_B0034 RBS
pSB1A2 AmpR
High
12bp
2091bp
Well 3O, Plate 1
--
A
G 21
BBa_J23039 Constitutive luxI
pSB1A2 AmpR
High
860bp
2939bp
Well 11K, Plate 3
140
A
W
G
23
BBa_I13504 GFP
pSB1A2 AmpR
High
875bp
2954bp
Well 16E, Plate 1
120/330
A
G
24
BBa_R0062 lux promoter
pSB1A2 AmpR
High
55bp
2134bp
Well 9G, Plate 1
90
A
G
25
BBa_J37032 GFP under lux promoter
pSB1A2 AmpR
High
938bp
3017bp
Well 8D, Plate 3
125/330
A
G
26
BBa_S03119 constitutive LuxR
pSB1A2 AmpR
High
998bp
3077bp
Well 19J, Plate 1
60
A
W
X 27
pSB1A3 pSB1A3 (blank)
pSB1A3 Amp
High
0bp
2157bp
Well 23E, Plate 1
--
W 30
---
T7 with GFP(strong)
---
Kan?
???
---
???
---
150
Our Parts
Physical information of our ligated parts:
A
W
G
S
ID
Name
Description
Plasmid
Resist
Copy
Size(B)
Size(P)
Stock
BBa_I719022 cI with GFP
pSB1A2
AmpR
High
933bp
3988bp
BBa_I791021
luxI coding region under ptet
pSB1A2
AmpR
High
860bp
2939bp
BBa_I791???
DsRed Express under ptet
pSB1A2
AmpR
High
Legend: A (Available), W (Working) [W - working in vivo , W - working in vitro , X - faulty], G (Checked on gel) [G - incorrect size; X - does not appear on gel], S (Sequenced)
Part Graveyard
General information on parts we do not like...
A
W
ID
Name
Description
Plasmid
Size
Status
Notes
A
W
01
BBa_E0040
GFP
pSB1A2
720bp
XL1, Mini
Redundant
A
03
BBa_J5528
pBad-GFP
pSB2K3
2093bp
failed-XL1
ara construct abandoned
A
04
BBa_J24677
plux-GFP
pSB1A2
1787bp
XL1, Mini
Wrong GFP ligated
A
06
BBa_I0500
pBad
pSB2K3
1210bp
XL1, Mini, Midi
ara construct abandoned
A
W
08
BBa_F1610
luxI
pSB1AK3
798bp
XL1, Mini, Midi
Substitute part J23039
A
W
09
BBa_R0010
lacI-plac
pSB1A2
200bp
XL1, Mini
Lac construct abandoned
A
W
10
BBa_E1010
RFP
pSB2K3
681bp
XL1, Mini
Substitute part I13507
A
12
BBa_S03550
araC
pSB1AC3
958bp
XL1, Mini, Midi
araC construct abandoned
W
14
BBa_R0051
pcI
pSB1A2
49bp
Biobrick
pcI construct abandoned
A
15
BBa_J07037
cI-GFP
pSB2K3
912bp
XL1, Mini
No RBS present on this part
A
W
16
BBa_E0240
GFP
pSB1A2
876bp
XL1, Midi
Redundant
W
20
---
AraC
pSB1AC3
1095bp
Planned
araC construct abandoned
A
W
28
BBa_P1010
pSB1A3 (ccd)
pSB1A3
0bp
failed-XL1
Strain not available
A
W
29
BBa_P1010
pSB1A2 (ccd)
pSB1A2
0bp
---
Strain not available
Construct Diagrams
The original (old) diagrams page was moved to old construct diagrams page. This page also contains the obsolete parts.
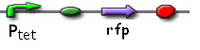
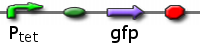
CBD Constructs
ptet GFP
Ptet promoting GFP
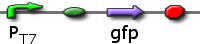
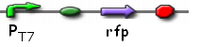
pT7 GFP
PT7 promoting GFP
ptet DsRed express
Ptet promoting dsRed Availability: Requires ligation
Promoter can be found in registry as: BBa_R0040
DsRed express is to be ordered from GeneArt
pT7 DsRed express
pT7 promoting dsRed Availability: Requires ligation
Promoter is being PCRed from plasmid derived externally
DsRed express is to be ordered from GeneArt
ID Constructs
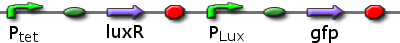
pLux GFP with LuxR
Ptet promoting LuxR, Plux promoting GFP
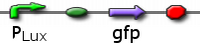
pLux GFP with LuxR
Plux promoting GFP
<<Two more constructs with DsRed express yet to be included>>