Biomod/2011/Harvard/HarvarDNAnos:Data&Output Final
<html> <head>
<style>
- column-one { display:none; width:0px;}
.container{background-color: #f5f5f5; margin-top:50px} .OWWNBcpCurrentDateFilled {display: none;}
- content {width: 0px; margin: 0 auto auto 0; padding: 1em 1em 1em 1em; align: center;}
- column-content {width: 0px; float: left; margin: 0 0 0 0;padding: 0;}
.firstHeading {display:none; width:0px;}
- globalWrapper{width:1280px; margin:auto}
body {background: #F0F0F0 !important;}
- column-one {display:none; width:0px;background-color: #f0f0f0;}
- content{border:none;margin: 0 0 0 0; padding: 1em 1em 1em 1em; position: center; width: 800px;background-color: #f0f0f0; }
.container{ width: 800px; margin: auto; background-color: #f0f0f0; text-align:justify; font-family: helvetica, arial, sans-serif; color:#f0f0f0; margin-top:25px; }
- bodyContent{ width: 1267px; align: center; background-color: #f0f0f0;}
- column-content{width: 1280px;background-color: #f0f0f0;}
.firstHeading { display:none;width:0px;background-color: #f0f0f0;}
- header{position: center; width: 800px;background-color: #f0f0f0;}
- footer{position: center; width:1280px;}
</style>
</head> </html>
<html>
</html>
Spherical Container Design
2011-06-20
- Sphere matching Han, et. al:
- An excel file containing all the staples for the sphere:
2011-07-06
Folding the Sphere
There are some mismatches here w.r.t Han's original staples, due to using a different sequence of the m13 clone.
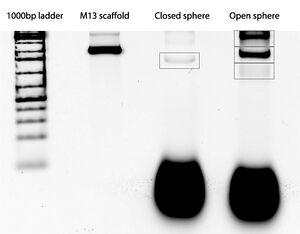
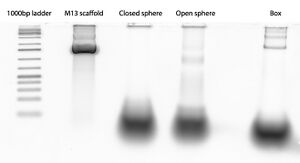
Gel
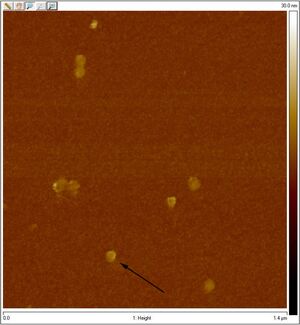
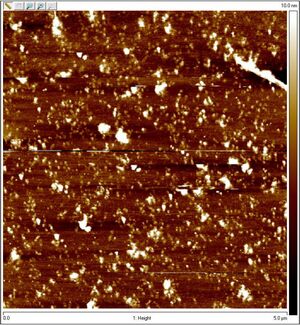
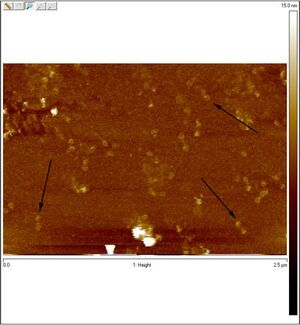
AFM
Unpurified closed spheres:
Unpurified open spheres:
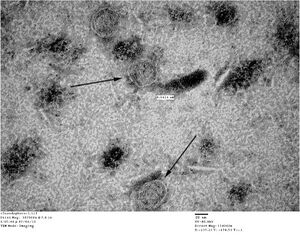
TEM
Unpurified closed spheres:
Unpurified open spheres:
2011-07-11
Gel:
Box Container Design
- evan needs to put data here
Gold Nanoparticles
Synthesis (unmodified)
2011-06-22
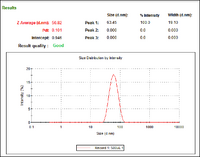
- 5nm gold nanoparticles ordered commercially (2 months old); there is aggregation in this sample
- DLS data for 5nm gold nanoparticles that we synthesized with Steve; there is no indication of aggregation
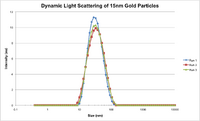
- DLS data for 15 nm gold nanoparticles that we synthesized with Steve; particles appear larger, but were a little warm when we ran the measurement, so it is possible that this data is slightly inaccurate
2011-06-23
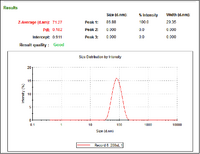
- Below are DLS outputs, which show that the more gold particles from a stock solution of 15nm particles introduced to a given volume of AuCl, the smaller the gold aggregates that form (more particles = wider distribution of the aqueous gold)
- Hydrodynamic Radius of Particles after Addition of 500 uL of 15nm Au Particle Stock Solution
- Hydrodynamic Radius of Particles after Addition of 200 uL of 15nm Au Particle Stock Solution
- Hydrodynamic Radius of Particles after Addition of 50 uL of 15nm Au Particle Stock Solution
- Hydrodynamic Radius of Particles after Addition of 20 uL of 15nm Au Particle Stock Solution
- Hydrodynamic Radius of Particles after Addition of 5 uL of 15nm Au Particle Stock Solution
2011-07-05
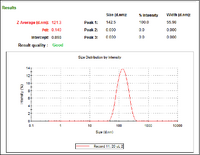
- The above files are links to the DLS measurements of my gold particles synthesized with a target diameter of 5nm before adding phosphene. The first measurement shows a uniform distribution of particles upon initial synthesis (with a hydrodynamic radius of about 12 nm). The second document, "PrePhosphene_4DaysLater" shows that there was a degree of aggregation of AuNP after sitting at 4 degrees C for four days without the addition of phosphene.
2011-07-18
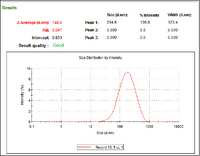
- The above documents are from gold particles synthesized with a target diameter of 5nm. As we can see from the first document, aggregates formed during initial synthesis. To remove these aggregates, I used a 2um vacuum filter. The second document shows the DLS measurement of the purified particles, clear of aggregates.
TEM Images and ImageJ size analysis
2011-07-19
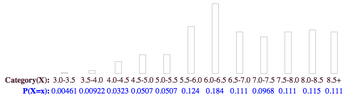
- Histogram showing the size distribution of our synthesized particles, with an average diameter of 6.61nm
2011-08-03
Post-Phosphene Modification
2011-07-05
- The above files are links to the DLS measurements of my samples after allowing my particles to finish the phosphene and salting protocols, respectively. The first measurement shows that the addition of phosphene stabilized the particles and prevented aggregation. The final measurement file, "PostSaltAddition" shows minimal aggregation of AuNP, but this is probably a result of using the centrifuge or effect of sodium ions allowing particles to come very close to each other (which is an inevitable result of the procedure).
2011-07-20
- The above document provides a DLS measurement of the stabilized particles after the phosphene addition protocol has been completed. There is no evidence of aggregation.
Conjugation with DNA
2011-07-11
- We attempted gel purification of our AuNP conjugated to DNA strands. Unfortunately, the gel we obtained appeared to only have a large smear. Ideally, we were hoping to see several distinct bands in our gel that indicated a discrete, successively increasing number of DNA strands attached to our gold nanoparticles.
2011-07-12
- After a second attempt to separate the conjugated particles via sucrose extraction we found a similar smear to yesterday. However, we decided to cut out the smear band and use Amicon filters to purify the fluid obtained (we also used the same filters to attempt purification of particles not introduced to the gel). We used a 1000MW cutoff filter, which unfortunately was effective. Luckily, we still have conjugate AuNP in a separate tube on the slow shaker. In the meantime though, I prepared an additional 200 uL of conjugated AuNP for use on our sphere and box designs.
- NOTE: As of July 18, we have reasoned that the cause of the smear and failure of the Amicon filter was that instead of coating our particles with 1 (or only a few) DNA strands, we had accidentally saturated the surface of our particles with DNA. We will proceed by adding far less DNA in the conjugation protocol, see here
Incorporation within DNA Origami
- Coming soon! (....so excited!)
Incorporation within Nanoparticle Chain
2011-06-23
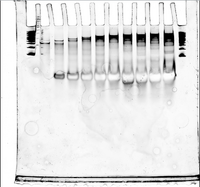
PAGE Gel of Handles+Ultramer Annealing Ladder
- 100 V, 10% TBE, 45 minutes
2011-06-24
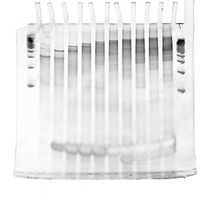
Second Attempt at PAGE Gel of Handles+Ultramer Annealing Ladder
- 100 V, 10% TBE, 2 hours
- still not enough separation, but clearly an upward trend indicating more and more handles attached to ultramer
- unclear what the white horizontal blotch near the bottom is; perhaps quenching due to too high concentration of SYBR-Gold because a lot of excess staples
PAGE Gel of Ultramer + All Nine Handles
- 100 V, 4 hours
- enough separation for extraction of top band, but should not have used SYBR-Gold
2011-06-28
PAGE Gel of Ultramer + All Nine Handles
- 100 V, 4 hours
- SYBR-Gold that was used to stain gel is faulty
2011-06-29
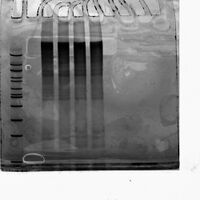
Overlay of Photo of Post-extraction Gel On Top of Gel Scan of Ultramer + Nine Handles
2011-07-14
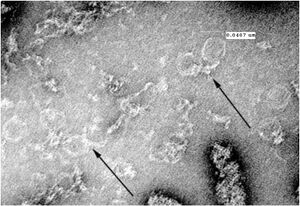
 TEM Image of Possible Nanoparticle Chains - Unstained Grid
TEM Image of Possible Nanoparticle Chains - Unstained Grid
 TEM Image of Possible Nanoparticle Chains - Stained Grid
TEM Image of Possible Nanoparticle Chains - Stained Grid
 TEM Image of Our Homemade 5 nm Gold Nanoparticles
TEM Image of Our Homemade 5 nm Gold Nanoparticles
Photocleavable Spacers
2011-07-25
Photocleavage Test 1: Using two strands with internal photocleavable spacers, directly from manufacturer-shipped tubes
- 10% TBE PAGE gel, 15 minute UV exposure for "cleaved" strands
Error creating thumbnail: An unknown error occurred. Error creating thumbnail: An unknown error occurred.
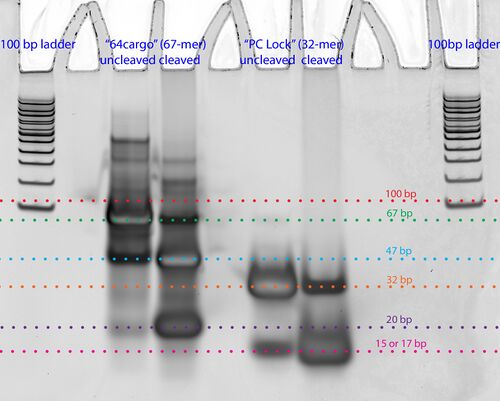
Photocleavage Test 2: Using gel-purified uncleaved strands from Photocleavage Test 1
- 10% TBE PAGE gel, 1 hour UV exposure for "cleaved" strands
SphereCAD
2011-06-21
Nick's excel file correlating caDNAno bp number with theta/phi position and whether the bp points into our out of the sphere: Media:CoordinatesSpreadsheet2.xlsx
2011-07-06