BME100 s2018:Group9 W0800 L4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | ||||||||||||||||||||||||||||||||
OUR TEAM
LAB 4 WRITE-UPProtocolMaterials
(hp://www.promega.com/resources/protocols/product‐informaon‐sheets/g/gotaq‐colorless ‐master‐mix‐m714‐protocol/)
ps: only use each only once. Never reuse disposable pipee ps. If you do, the samples will become cross‐contaminated
OpenPCR program
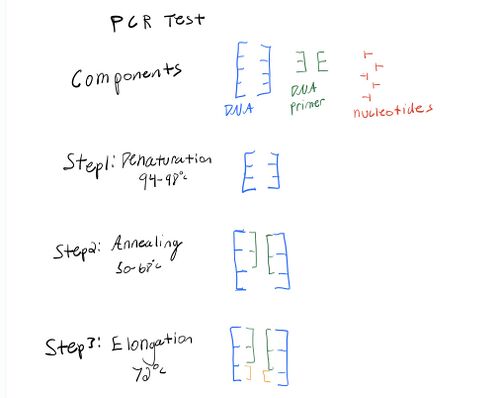
Research and DevelopmentPCR - The Underlying Technology What is the function of each component of a PCR function? The purpose of the polymerase chain reaction is to copy a region of DNA multiple times in a test tube rather than an actual organism. That way it can eventually be analyzed in a lab. Understanding what the following functions are capable of, enhances the understanding of PCR. Template DNA is used as a master copy for replication in the lab. Primers are the nucleotides that allow DNA synthesis to start. Primers are pieces of single strand DNA. In order to start the process of DNA synthesis, the two primers attach themselves to the template DNA. Taq Polymerase is an enzyme that makes new strands of DNA. Enzymes are typically controlled by heat, which is why taq polymerase separates the strands of DNA very quickly. Deoxyyribonucleotides (T,G,C)are the building blocks of DNA during the PCR function. Mainly these nucleotides generate gene sequences which means the nucleotides match to make a "sequence".
What happens to the components during each step of thermal cycling? Thermal cycling consists of steps that make DNA replicate into billions of copies. The intial step is where the DNA is heated up and causes the DNA strands to split; also providing access to the primers. During the second step (denature)an excess amount of primers are added. The reaction mixture must be cooled that way double strands of DNA are able to form again and they will always bind to primers. During the third and fourth step (Anneal, extension), a mixture containing deocyribosnucleotides, will be combined with an enzyme called Taq polymerase. The enzyme matches up the nucleotides during this step, in order to make the DNA sequence. The Final step to the polymerase reaction is simply replicating the exact same steps above multiple times in order to make billions of copies of the region of the DNA template. Base-pairing. driven by hydrogen bonding, allows base pairs to stick together. Which base anneals to each listed? Adenine: Thymine: Cytosine: Guanine: Adenine pairs with Thymine; Cytosine pairs with Guanine.
Base-pairing occurs during the annealing and extension steps. During step two(annealing), when the temperature of the reaction solution is cooled to 57 degrees Celsius, two primers attach to the strands of single strand of DNA. The primers bind to the DNA by base-pairing and form gene sequences. During the fourth step(extension), when the reaction solution is heated to 72 degrees Celcius, Taq Polymerase combines with the deoxyribonucleotides through base-pairing in order to make the final DNA sequence.
SNP Information & Primer DesignBackground: About the Disease SNP Single nucleotide polymorphism is a difference in a nucleotide. For example, the SNP might replace the nucleotide Cytosine with the nucleotide Thymine in a certain region of DNA. SNP's are found mainly within a certain part of DNA between genes.One of the reasons why SNP's are so important is because they can ac as bio-markers and help scientists locate what gene is associated with a disease; potentially affecting the genes function. These changes in the genome can also determine how a human reacts to various types of drugs, viruses, bacteria and other substances. Citation: https://www.sciencedirect.com/topics/neuroscience/single-nucleotide-polymorphism Primer Design and Testing Part 1 Identifying vocabulary: Polymorphism is the difference between genes; it is what creates different different individuals amongst a type of species. Nucleotides are the building blocks to DNA,the main bases are Adenine, Thymine, Guanine and Cytosine. Nucleotides are made up of three main components, the nitrogen bases, five-carbon sugar and a phosphate group. The species rs1044498 is located amongst the homo-sapien species. The chromosome it is located specifically on is chromosome 6:131851228. The clinical significance of this SNP is listed as other. The conditions linked to this particular SNP are bone disorders in type two diabetes. Specificaly females who were diagnosed with bone mineral density and classified by their diabetes status. Part 2
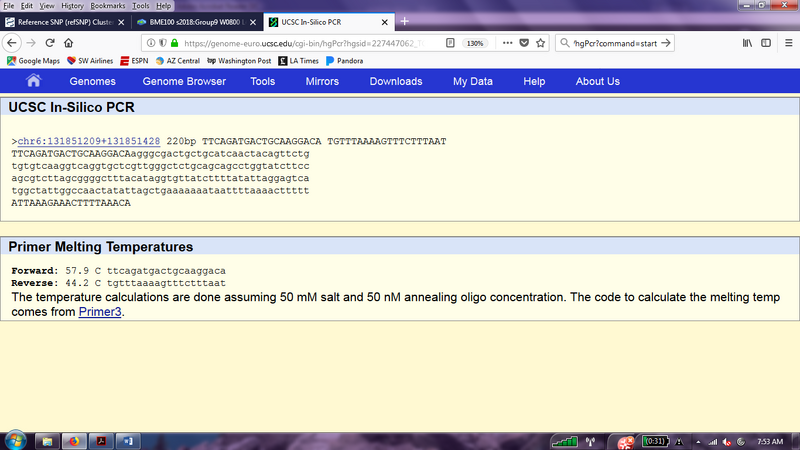
Part 3 The results of the primer testing in UCSC In-Silico PCR, validated that the primers corresponded to the exact mutated genes. The first picture represents the results of the primers matching that of the mutation on the gene. In other words, the primers are located on chromosome 6, 220 base pairs apart(200 base-pairs long and 20 bases long).The forward non-disease primer is 5'TTCAGATGACTGCAAGGACA 3' and the reverse non-disease primer is 5'TGTTTAAAAGTTTCTTTAAT3'. The disease froward primer is 5'TTC AGATGACTGCAAGGACC3'. The disease reverse primer is 5'TGTTTAAAAGTTTCTTTAAT3'.
| ||||||||||||||||||||||||||||||||