BME100 s2018:Group8 W1030 L4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
OUR TEAM
LAB 4 WRITE-UPProtocolMaterials
PCR Reaction Sample ListTable 1: Patient Information
DNA Sample Set-up Procedure
Repeat Steps 1-8 for tubes G8-,G8 1-1, G8 1-2, etc. OpenPCR programTable 2: Summary of the temperature and time for each step in the procedure of PCR.
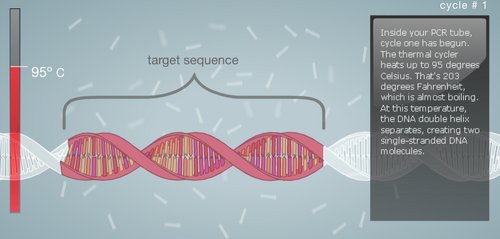
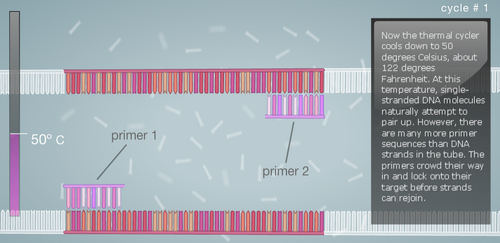
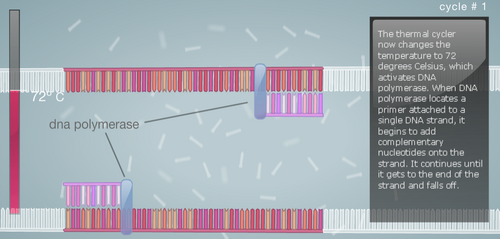
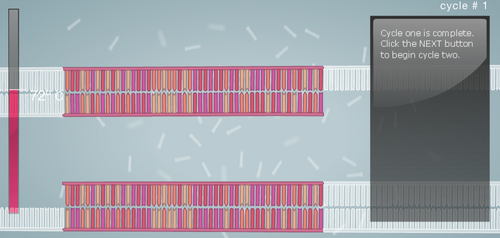
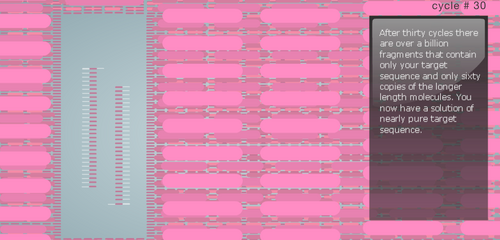
Research and DevelopmentPCR - The Underlying TechnologyFunction of Each Component in a PCR Reaction First let's look at the template DNA, which is the DNA extracted from the hair, saliva, or blood of a human being. With a tiny portion of DNA extracted from the template we can generate billions of copies in about 30 cycles inside the thermal cycler. Next are the primers, these are custom built of any nucleotide sequence that we like. Their function is to attach to the top end and the bottom end of the DNA strands through complementary base pairing. After the primers, we have the Taq Polymerase, a thermostable DNA polymerase, this is because it is derived from the bacteria called Thermus aquaticus, found in hot springs and function efficiently at boiling temperatures. Once the DNA polymerase identifies the location of the primers, its function is to generate a copy of cell's DNA from the nucleotides present around. Finally, the Deoxyribonucleotide, which are the sub-units of DNA. They are made up of a nitrogenous base, a phosphate, and a deoxyribose sugar. The nitrogenous bases include Adenine (A), Thymine (T), Guanine (G), Cytosine (C), and each base is attached to the other two components. The base pairs also attach with their complements to give us a a two stranded DNA; A-T and G-C. A Breakdown of each temperature in the cycle For each cycle in the PCR we follow the same temperature cycle. The DNA Thermal Cycler starts at 95°C, this causes the double helix to separate and gives us single strands. Then the temperature goes down to 50°C, and this causes the strands to connect with their counterpart. However, because of the abundance of primers, the strands connect with them instead. At 72°C, the DNA polymerase is activated, and it begins adding the nucleotides once it locates the primer. The DNA polymerase goes till the end, and then falls. This same cycle of temperature if continued 25 times, and by the end we have over a million copies of the DNA. Following is a step by step visual representation of what happens at each step in the DNA Thermal Cycler. Image 1: At 95°C Image 2: At 50°C Image 3: At 72°C Image 4: End of Cycle #1 Image 5: After 30 cycles Source: http://learn.genetics.utah.edu/content/labs/pcr/ With the help of hydrogen bonding, the base pairs are able to stick together. As mentioned earlier: Adenine complements Thymine, and Guanine complement Cytosine. In PCR, we observe base pairing twice. First, when the primer attaches itself to each strand, and it is able to do so because the nitrogenous bases find their pairs in the DNA strands. Second, when DNA polymerase combines the DNA strands with their complementing base pairs.
SNP Information & Primer DesignBackground: About the Disease SNPAdenine, cytosine, guanine, thymine are all examples of nucleotides and are the basic units of DNA. Within those strands of DNA, polymorphism can occur. Polymorphism is an occurence of different forms within a section of nucleic acid than what is normally found. In SNP or Single Nucleotide Polymorphism, a single nucleotide will contain a varied portion within a DNA sequence. SNP rs1044498 is found primarily on chromosome 6 in position 13851228 in homo sapiens(humans) and has been studied by many. It has been found to be connected with type two diabetes, insulin blocking and causes problems with your bones (bone mineralization). Studies suggested ENPP1 (ectonucleotide pyrophosphatase/phosphodiesterase 1) resists insulin and inhibits bone mineralization. It also is connected to ATP binding, calcium ion binding and 3’-phosphoadenosine 5’-phosphosulfate binding. You can find alleles(genes with an alternate form) within the same location on chromosome 6. The codon for the non-disease allele for SNP is AAG. This changes to CAG because of SNP. Primer Design and Testing
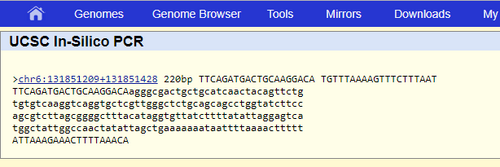
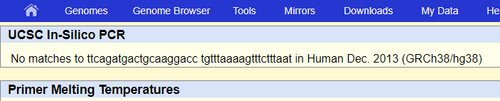
The primer test we completed shows where primers are located. This process showed that the primers were 220 base pairs apart and on the 6 chromosome. The same exact data we found in the primer design. They were 200 base pairs away from each other and were 20 bases long. When we tested the disease primers we found that there were no matches. This is correct because it has the SNP mutation, which is not considered a healthy primer that exists in the human genome. Non-Disease Primer: Disease Primer:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||