BME100 s2018:Group7 W1030 L4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | ||||||||||||||||||||||||||||||||
OUR TEAM
LAB 4 WRITE-UPProtocolMaterials
HEATED LID: 100°C INITIAL STEP: 95°C for 2 minutes NUMBER OF CYCLES: 25
FINAL STEP: 72°C for 2 minutes FINAL HOLD: 4°C
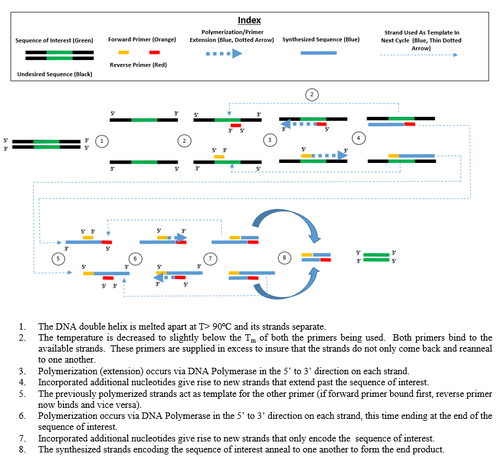
Research and DevelopmentPCR - The Underlying Technology Function of Each Component of a PCR Reaction Template DNA replicates itself into 100 billion identical copies.Primers attach to sites on the DNA strands that are at either end of the segment that is being copied. They are important for replicating specific DNA sequences since there is little chance of coming in contact with the wrong sites. Taq Polymerase are polymerase molecules that act like tiny machines that read the DNA code and then attach matching nucleotides to create DNA copies. They are elongated. The deoxyribonucleotides (dNTP’s) have to be added in order to elongate the nucleotides.The DNA polymerase grabs nucleotides that are floating around in the liquid around it and attaches them to the end of a primer.
At 95 degrees celsius, the DNA double helix begins to separate, which then creates two single-stranded DNA molecules. The thermal cycler cools down to 50 degrees Celsius, causing the single strand DNA molecules to pair up but there are too many primer sequences than DNA strands. The primers overcrowd and lock onto their target before strands can rejoin. The thermal cycler warms up to 72 degrees celsius, activating the DNA polymerase. When DNA polymerase locates an attached primer with a single DNA strand, it begins to add a complementary nucleotide. It continues until the end of the strad and then falls off. Two strands with primer one and primer two appear.
A-T and G-C are base pairs. They are complementary nucleotides. Base Pairs in Thermal Cycling When the temperature dropped, the single strand DNA molecules to pair up but there are too many primer sequences than DNA strands. The primers overcrowd and lock onto their target before strands can rejoin. This causes the pairing of nucelotides. When the temperature increases, the DNA polymerase locates an attached primer with a single DNA strand, it begins to add a complementary nucleotide. It continues until the end of the strad and then falls off.
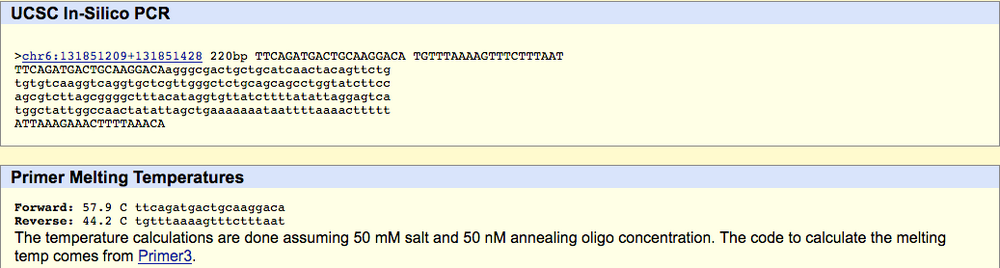
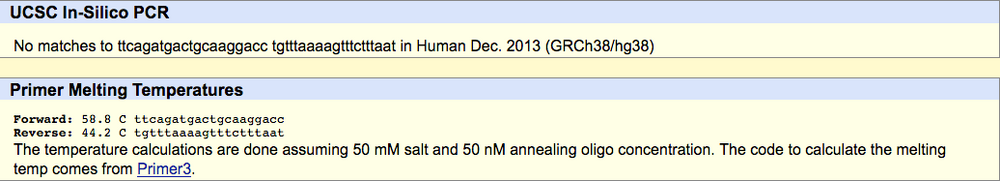
SNP Information & Primer DesignBackground: About the Disease SNP SNP, also known as single nucleotide polymorphism, is when a base pair in a strand a DNA undergoes variation. The variation that was discovered in this lab was found on the ectonucleotide pyrophosphatase/phosphodiesterase 1 (ENPP1) gene which was found on the 6:131851228 chromosome. ENPP1 usual functions involve protein homodimerization activity, NADH pyrophosphatase activity, and nucleoside-triphosphate diphosphatase activity. A variation in this location results in the onset of Type 2 Diabetes in homo sapiens. The SNP of this disease changes alleles found at the position 131851228 which changes AAG to CAG which results in the disease. Primer Design and Testing The first primer test involved the non-disease forward primer, 5'-TTCAGATGACTGCAAGGACA-3', and the non-disease reverse primer, 5'-TGTTTAAAGTTTCTTTAAT-3'. When put into In-Silico PCR, the result was 220 bp. The forward primer's melting temperature was 57.9°C, and the reverse primer's melting temperature was 44.2°C. The second primer test involved the disease forward primer, 5'-TTCAGATGACTGCAAGGACC-3', and the non-disease forward primer, 5'-TGTTTAAAAGTTTCTTTAAT-3'. When put into In-Silico PCR, there were no matches for the corresponding primers. the disease forward primer had a melting temperature of 58.8°C and the reverse primer had a melting temperature of 44.2°C.
ReferencesENPP1 Ectonucleotide Pyrophosphatase/Phosphodiesterase 1 [Homo Sapiens (Human)] - Gene - NCBI. https://www-ncbi-nlm-nih-gov.ezproxy1.lib.asu.edu/gene?cmd=Retrieve&dopt=Graphics&list_uids=5167#gene-ontology. Accessed 14 Mar. 2018. How Jack Black Channeled His Inner Teenage Girl For Jumanji. 18 Aug. 2017, https://www.cinemablend.com/news/1693510/how-jack-black-channeled-his-inner-teenage-girl-for-jumanji. Hsiao, Tun-Jen, and Eugene Lin. “The ENPP1 K121Q Polymorphism Is Associated with Type 2 Diabetes and Related Metabolic Phenotypes in a Taiwanese Population.” Molecular and Cellular Endocrinology, vol. 433, 15 2016, pp. 20–25, doi:10.1016/j.mce.2016.05.020. Karen Gillan Speaks out after Fans Attack Her Skimpy Attire in Jumanji Reboot | Daily Mail Online. http://www.dailymail.co.uk/tvshowbiz/article-3801526/Karen-Gillan-speaks-fans-attack-skimpy-attire-Jumanji-reboot.html. Accessed 14 Mar. 2018. Kasdan, Jake. Jumanji: Welcome to the Jungle. 2017, http://www.imdb.com/title/tt2283362/. Neamati, Nahid, et al. “The ENPP1 K121Q Polymorphism Modulates Developing of Bone Disorders in Type 2 Diabetes: A Cross Sectional Study.” Gene, vol. 637, Dec. 2017, pp. 100–07, doi:10.1016/j.gene.2017.09.042. “Polymerase Chain Reaction.” Wikipedia, 12 Mar. 2018, https://en.wikipedia.org/w/index.php?title=Polymerase_chain_reaction&oldid=830087355. Reference SNP (RefSNP) Cluster Report: Rs1044498 ** With Other Allele **. https://www-ncbi-nlm-nih-gov.ezproxy1.lib.asu.edu/projects/SNP/snp_ref.cgi?rs=1044498. Accessed 14 Mar. 2018. Rs1044498 - SNP - NCBI. https://www-ncbi-nlm-nih-gov.ezproxy1.lib.asu.edu/snp/?term=rs1044498. Accessed 14 Mar. 2018. Severson, Tyler J., et al. “Genetic Factors That Affect Nonalcoholic Fatty Liver Disease: A Systematic Clinical Review.” World Journal of Gastroenterology, vol. 22, no. 29, Aug. 2016, pp. 6742–56, doi:10.3748/wjg.v22.i29.6742. The Classic Robin Williams Movie Jumanji’s Kevin Hart Would Like To Work On Next. 15 Dec. 2017, https://www.cinemablend.com/news/1744820/the-classic-robin-williams-movie-jumanjis-kevin-hart-would-like-to-work-on-next. UCSC In-Silico PCR. http://www.genome.ucsc.edu/cgi-bin/hgPcr?command=start. Accessed 14 Mar. 2018. | ||||||||||||||||||||||||||||||||