BME100 s2018:Group6 W1030 L4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | ||||||||||||||||||||||||||||||||
OUR TEAM
LAB 4 WRITE-UPProtocolMaterials
OpenPCR program The thermal cycler's program, referred to here as the OpenPCR program, will run a cycle of heating and cooling on the extracted DNA and allow the PCR reaction to occur. This program will begin with an initial heating at 95°C for 2 minutes. After the sample has been heated, the program will alter the temperature between 95°C for 30 seconds, 57°C for 30 seconds, and 72°C for 30 seconds. This will respectively denature, anneal, and then extend the DNA into two separate, cut strands. After twenty five cycles of these three temperature changes, the mixture will be held at 72°C for 2 minutes, then removed.
Research and DevelopmentPCR - The Underlying Technology The Components of PCR PCR amplification is a very powerful and helpful tool for human diagnostics, environmental monitoring, and scientific research. The human genome contains 3 billion base pairs which makes it very difficult to evaluate (OPEN PCR). PCR is a technique to make copies of a certain section of DNA. The key components of a PCR reaction are: template DNA, primers, Taq Polymerase, and nucleotides. The template DNA is the specific region of DNA intended to be replicated for further study. When completing the reaction, there are 2 primers for the DNA piece. These primers are a short sequence of nucleotides that give a starting point for DNA synthesis. (Khan Academy) The primers attach to the sites at either end of the DNA strand and are crucial pieces in targeting the proper sites. Then the deoxyribonucleotides or dNTP’s. These are the nucleotides that are the building blocks of DNA and their purpose is to make up the DNA copies. After the primers flank the target sites, Taq polymerase comes into play. Taq polymerase is the DNA polymerase that starts the replication process and synthesizes the new strands of DNA. Thermal Cycling During the thermal cycler, three clear-cut changes occur at the various temperatures. Firstly, the two minutes of heating before it begins cycling, it is held at 95°C to completely denature all DNA held in the cycler. Then the cycle begins. Held at 95°C, the DNA is denatured, so that it separates from a double-stranded molecule into two single strands of DNA. It is then cooled to 57°C, which is a perfect temperature to encourage pairing. Since there are dNTPs added to the reaction mix, they will lock more readily to the single DNA strands, crowding out the previously-severed partner strand. Finally, the mixture is raised to 72°C, which allows Taq DNA polymerase to bind these dNTP primer sequences to the single strand of DNA to which they are locked. This will create two new double stranded DNA molecules for every one molecule that began the cycle. It then held at 72°C for two minutes to ensure that all of the targeted sections are bound and that the DNA is correctly formed back into its double helix. It is held at 4°C to cool the samples down before they are removed. Base Pairing The nucleotides are specifically paired. Adenine pairs with thymine and cytosine pairs with guanine. In thermal cycling, the base pairing occurs during the anneal and extend steps of the process. During annealing, the DNA is cooled to about 50°C so that the primers are able to bind to their complementary sequences on the DNA strands. Then during the extension, the Taq polymerase extends the primers by adding the nucleotides by base-pairing.
SNP Information & Primer DesignBackground: About the Disease SNP Nucleotides are a compound of a nucleoside linked to a phosphate group. That forms the basic structure of nucleic acids, like DNA. The occurrence of different forms of members within a population is known as polymorphism. [[1]] Therefore, a Single Nucleotide Polymorphism, SNP, rs1044498 is found in homo sapiens, humans. SNPs are a common form of genetic mutation that occur when there is variance with a single nucleotide, within a DNA sequence. SNP rs1044498 is found on chromosome 6 position 13851228. [[2]] This SNP is associated with development of bone disorders in type two diabetes. [[3]] The clinical significance of this SNP is pathogenic. The function ectonucleotide pyrophosphatase/phosphodiesterase1, ENPP1, protein is to control cell response, such as to Zinc ion binding, scavenger receptor activity, and insulin receptor binding, along with many others. [[4]] Alleles are genes that are of an alternate form (one of two or more). which arise by a mutation that is found at the same location on a chromosome. The codon for the non-disease allele from the given SNP, is AAG. Which changed to CAG due to the allele associated with the diseased SNP.
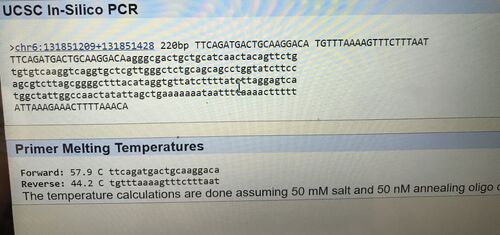
The primer test for the non-disease forward and reverse primers was successful because the result from the USCS In-Silico PCR web-page gave a 220 bh sequence from the chromosome. The SNP was positioned at 131,851,228 and 200 base pairs to the right of the disease SNP was 131,851,428. The non-disease forward primer was recorded to be TTCAGATGACTGCAAGGACA and the non-disease reverse was TGTTTAAAAGTTTCTTTAAT. When the disease forward and reverse primers were inputted into the web-page, it resulted in no matches. The disease forward primer was the same as the non-disease forward primer, but the last base was changed to be identical to the disease SNP nucleotide (from A to C).The disease reverse primer was kept the same as the non-disease reverse primer. The reason the disease forward and reverse primers resulted in no matches is because it is not the healthy primer, so it will show up as no matches because it is different from the healthy primer. Sources
| ||||||||||||||||||||||||||||||||