BME100 s2018:Group5 W1030 L6
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | |||||
OUR COMPANY
Our Brand Name LAB 6 WRITE-UPBayesian StatisticsOverview of the Original Diagnosis System The number of four-person teams diagnosing the 14 patients was 7, with two patients per team. In order to prevent error, three PCR tests were done per patient and three images were taken and analyzed per tests. A positive and negative control tests were performed (with three images taken of each). Also, Three images of a no-concentration solution (water droplet) were taken as a controls for imageJ results. The number of conclusive results was 9 out of 14, and the remaining 5 were inconclusive.
In calculation 2, the probability that a patient will have a negative diagnostic signal given a negative final test conclusion is about 80%. The probability that a patient will get a negative final test conclusion given a negative diagnostic signal is also about 80%. This implies that the individual PCR replicates for concluding that a person has the disease or not is more reliable when both are negative.
In calculation 3, the probability that a patient will have a positive test conclusion given the patient develops the disease is close to 50%. The probability that a patient develops the disease, given a positive test conclusion is more than 100%. This implies that the PCR is reliable in predicting the disease from a positive test conclusion but is not reliable in diagnosing the disease once it is already present. In calculation 4, the probability that a patient will get a negative final test conduction given the patient did not develop the disease is around 80%. The probability that a patient will not develop the disease given a negative final test conclusion is more than 100%. This implies that the PCR is reliable in predicting the disease from a negative test conclusion but is only partially reliable in diagnosing the disease once it is already present.
Intro to Computer-Aided Design3D Modeling
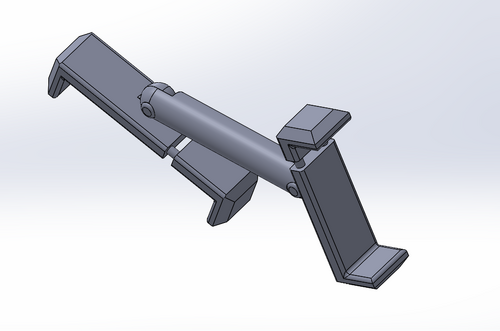
In the original fluorimeter design, the distance between the phone camera and the LED light/ drop to be imaged could vary wildly between pictures. Each time the phone is picked up and replaced between trials, its position could be different from the last trial. This causes errors in imaging and image analysis because the distance affects the intensity of green light detected by the phone camera. Because this is the most important part of the imaging step in this lab, it is very important to minimize the possible error in this step.
Feature 1: ConsumablesA "very important" component of an experiment means that these ingredients/items are vital to the experiment. Essentially, the experiment could not be constructed nor conclude to accurate results without these consumables. If in fact, we did redesign the PCR machine with shorter more efficient tubes, not many accommodations would have to be made in relevance to the consumables included. Nonetheless, our kit would include everything needed to complete the PCR reaction as well as the fluorimeter function. This would include the PCR mix, the SYBR Green Solution, and the primer and buffer mixes. Along with the vital chemical components needed to complete the PCR reaction, our kit will also include disposable pipette tips and glass slides. Through our kit, the consumer will be covered through RegEnergy with all of the consumable aspects of the experiment, and will only be responsible for already being equipped with a micropipette and lab protective gear, such as lab coats and gloves. And of course, the RegEnergy PCR Kit we will include a standard PCR machine and our edited Fluorimeter.
Feature 2: Hardware - PCR Machine & FluorimeterWe will not alter any parts of the PCR machine in our design. As it is, the Open PCR machine has may advantages, including being open source, being fairly inexpensive, having a clear and straightforward interface, and being easy to use. In this lab, we did not conclude that any errors or inaccuracies were due to this machine or this step in the lab. For these reasons, we will keep the Open PCR machine in our redesign without changes.
| |||||