BME100 s2018:Group5 W1030 L4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | ||||||||||||||||||||||||||||||||
Group 5
LAB 4 WRITE-UPProtocolMaterials
2. Cut empty PCR tubes so they will fit into the OpenPCR machine 3. Label the sides of each tube with labels listed in above table 4. Place tubes in rack 5. Starting with positive control tube, properly pipette 50 μL of PCR reaction mix into the tube. DISCARD TIP! 6. To this same tube, add the positive control DNA/ primer mix. This tube now contains 100 μL. 7. Repeat steps 5 and 6 for the remaining test tubes. Be sure to use the correct DNA/ primer mix for each reaction tube. 8. Be sure all reaction tube lids are tightly closed. 9. Place tubes into PCR machine. 10. When all slots are filled (multiple groups use the same machine) it is ok to start the run.
INITIAL STEP: 95°C for 2 minutes NUMBER OF CYCLES: 25 Denature at 95°C for 30 seconds, Anneal at 57°C for 30 seconds, and Extend at 72°C for 30 seconds FINAL STEP: 72°C for 2 minute
Research and DevelopmentPCR - The Underlying Technology
There are many components that work together to perform PCR reactions. The first of those many components being Template DNA, which is sample DNA that contains the target sequence. The second component of the reaction comes from the Primers, which is the function is to bind to sequences that flank the region of interest in the template DNA. Powerful tools for copying DNA. The third is the Taq Polymerase which high temperatures binds nucleotides to a DNA template, copying the DNA. The last and arguable most important part(s) are the Deoxyribonucleotides(dNTP’s), which supply the bricks/building blocks during PCR. It strings together dATP, dTTP,dGTP, and dCTP to produce DNA.
During thermal cycling these different components undergo various changes. In the initial step the DNA begins in double helix form before reaching 95 degrees Celsius. At the Denature Stage which occurs right around 95 degrees celsius, the double stranded DNA template denatures and becomes single stranded. Then at the Anneal Stage at around 57 degrees Celsius, that piece is cooled down to allow the nucleotide primers to base pair with the DNA template. Following that, the Extend Stage occurs at a little warmer temperature of 72 degrees and it is warmed up to allow the Taq-DNA to perform synthesis. The Taq releases loose nucleotides along the the DNA template so that the newly made DNA is extended from the primer. After that, 1 whole cycle has elapsed and the copying continues. After three cycles the desired fragments start to appear. Two strands that begin with primer one and end with primer two. After cycle 4, will have 8 of the desired fragments. After 30 cycles, more than 1 billion fragments of the desired fragments exist.
Base-pairing occurs during the anneal and extend stage within thermal cycling. During anneal the nucleotide primers to base pair with the DNA template and during extend the solution is Warmed up to allow the Taq-DNA to perform synthesis. The Taq releases loose nucleotides along the the DNA template so that the newly made DNA is extended from the primer.
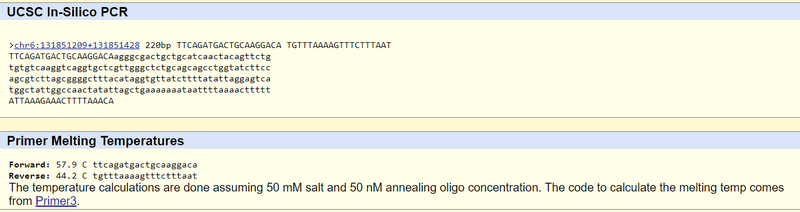
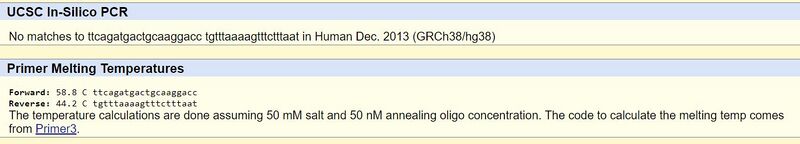
SNP Information & Primer DesignBackground: About the Disease SNP SNP stands for single nucleotide polymorphism. A phenotypic change as a result of a deletion, insertion, or switch of a nucleotide base pair. The SNP variation rs1044498 is found in Homo sapiens and causes bone disorders in Type 2 diabetes. The variation is located of the 6:131851228 chromosome. This disorder is more formally known as osteoporosis. Primer Design and Testing
The non-disease forward primer was identified by counting 19 nucleotides to the left of the base pair in question and recording these left to right (20 total). The non-disease reverse primer was identified by counting 200 nucleotides to the right of the base pair in question and recording 20 nucleotides reading right to left on the reverse strand starting with the nucleotide that was 200 to the right of the base pair in question. The disease primer strands were identified by replacing the last nucleotide (A) of the forward primer with the disease codon's switched nucleotide (C) and leaving the rest. Both the disease primers and non-disease primers were tested in the UCSC In‐Silico PCR website. Below are images of the results.
| ||||||||||||||||||||||||||||||||