BME100 s2018:Group5 W0800 L4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | ||||||||||||||||||||||||||||||||
OUR TEAM
LAB 4 WRITE-UPProtocolMaterials
OpenPCR program
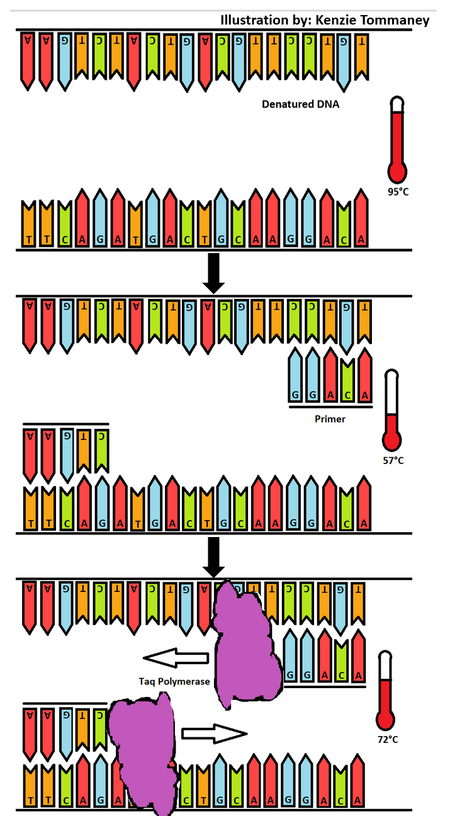
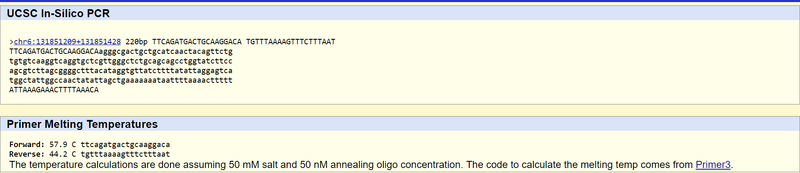
Research and DevelopmentPCR - The Underlying Technology What is the function of each component of a PCR reaction? In a PCR or a Polymerase Chain Reaction, there are 4 main components that include the template DNA, primers, taq polymerase and deoxyribonucleotides or dNTP's. The DNA template is the DNA that we want to replicate using the PCR reaction. It is usually placed within a PCR tube and kept cool before being placed in the thermocycler. The primers, usually two used in PCR are the components that serve as the starting point of the DNA replication before taq polymerase can come in and replicate the DNA. Taq polymerase is the enzyme that is responsible for copying and creating the DNA from the template strand in order to create a new identical strand of DNA. The dNTP's are the nucleotide bases that will make up the new copy of the DNA strand. For instance the dNTP's include dATP, dTTP, dCTP and dGTP. What happens to the components during each step of thermal cycling? Thermal cycling is one of the main processes of the PCR reaction. It includes 6 steps that requires the DNA in the PCR tube to be heated at 95°C for 2 minutes, denatured at 95°C for 30 seconds, annealed at 57°C for 30 seconds, extended at 72°C for 30 seconds, kept at 72°C for 2 minutes and then finally held at 4°C. In the initial step all of the materials including the template, two primers, taq polymerase and dNTP's are heated from room temperature to 95°C. At this point the template strand is denatured into two strands of DNA molecules. Next, in the annealing step the temperature drops to 57°C so that the two primers can each attach to the two single strands of DNA. Then in the the extending step of the thermal cycling, the temperature goes back up to 72°C in order for the taq polymerase to attach to the two primers and attach dNTP's to the single template strands of DNA. ATGC DNA is made up of four types of nucleotides designated by the letters A, T, C and G. Adenine and guanine are classified as purines while cytosine and thymine are classified as pyrimidines. These classifications are due to the structure of the nitrogen bases and phosphate groups of the nucleotides. Adenine and thymine are two bases that pair together and cytosine and guanine are another pair. Hydrogen bonding is the reason that these base pairs the purines and pyrimidines can stick together to form DNA. During which two steps of thermal cycling does base pairing occur? During thermal cycling the two steps where base pairing occur are during annealing and extending. The temperature drops to 57°C which is what allows the two primers to attach themselves to the DNA strand. SNP Information & Primer DesignBackground: About the Disease SNP For this lab, we will be investigating a disease single nucleotide polymorphism, or SNP. A nucleotide is a basic building block of DNA or RNA made from a base, sugar and phosphoric acid. The base for DNA can include adenine, thymine, guanine, and cytosine and these are placed in order to create a DNA sequence. When a nucleotide in a genome sequence is changed, it can create a polymorphism, or a genetic variation in the DNA of a population that is relatively common. The SNP we are testing for, rs1044498, is found in Homo sapiens and is located in the 6:131851228 chromosome. For clinical significance, it is considered benign. However, it could be a risk factor for obesity, diabetes mellitus type 2, arterial calcification of infancy, susceptibility to insulin resistance, and hypophosphatemic rickets. The SNP was closely associated with the bone disorders osteoporosis and osteopenia, especially when the patient has diabetes. It is recommended to check for this variation in patients with type 2 diabetes to predict these bone disorders. This variation is found in the DNA sequence ENPP1, which stands for ectonucleotide pyrophosphatase/phosphodiesterase 1. Its main functions are 3’-phosphoadenosine 5’-phosphosulfate binding, ATP binding, and NADH pyrophosphatase activity. The disease SNP forms two alleles, which are one of several versions of a gene, which arise due to mutation at a specific spot on the chromosome. The non-disease allele contains the codon AAG, while the disease-associated allele contains the codon CAG. The numerical position of the SNP is 131851228. Primer Design and Testing The non-disease forward primer is 5’-TTCAGATGACTGCAAGGACA-3’ The other primer will be 200 bases to the right of the SNP, so at position 131851428. The non-disease reverse primer is 5’-TGTTTAAAAGTTTCTTTAAT-3’ The disease forward primer is 5’-TTCAGATGACTGCAAGGACC-3’ The disease reverse primer is 5’-TGTTTAAAAGTTTCTTTAAT-3’
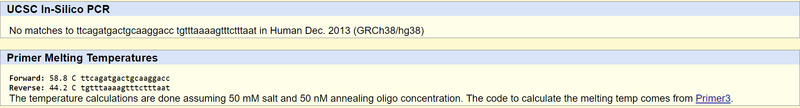
Test results for disease primers: These results match what was expected. The disease primers yielded no results because the website used a non-disease human genome sequence. | ||||||||||||||||||||||||||||||||