BME100 s2018:Group4 W1030 L6
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
OUR COMPANY
Our Brand Name LAB 6 WRITE-UPBayesian StatisticsOverview of the Original Diagnosis System BME 100 tested patients for the disease-associated SNP by completing PCR reactions and evaluating the results using a fluorimeter and a smartphone. The division of labor in which completed this testing was 8 teams composed of 4 students each diagnosed 14 total patients. Steps were taken to prevent error, such as three trials were completed for each patient's DNA, positive and negative controls were also tested three times. Furthermore, for each of these separate trials, three images were taken to analyze in Image J. Additionally, concentrations of DNA and water were tested to further study the differences. All of the data was compiled into a spreadsheet to highlight the results of the PCR reactions for the patients. The spreadsheet had many successful conclusions and inconclusive results, as well as black data. The inconclusive results were due to controls or results that did not match the data needed to diagnose the patient. This was likely because of a wide array of human and experimental lab. The major challenge that our group faced while completing this lab was inconsistencies with the angle and distance of the smartphone due to inadequate equipment, and this led to inconsistencies within the results on Image J. This challenge definitely affected our results as the images were inconsistent and thus hard to compare with each other.
P(A|B) = P(B|A) *P(A) / P(B) Calculation 1: What is the probability that a patient will get a positive final test conclusion, given a positive PCR reaction?
Calculation 2: What is the probability that a patient will get a negative final test conclusion, given a negative diagnostic signal?
Calculation 3: What is the probability that a patient will develop the disease, given a positive final conclusion?
Calculation 4: What is the probability that a patient will not develop the disease, given a negative test conclusion?
Which calculation describes the sensitivity of the system regarding the ability to detect the disease SNP? Calculation 1, 0.54 Which calculation describes the sensitivity of the system regarding the ability to predict the disease? Calculation 3, 1.15 Which calculation describes the specificity of the system regarding the ability to detect the disease SNP? Calculation 2, 0.72 Which calculation describes the specificity of the system regarding the ability to predict the disease? Calculation 4, 1.10
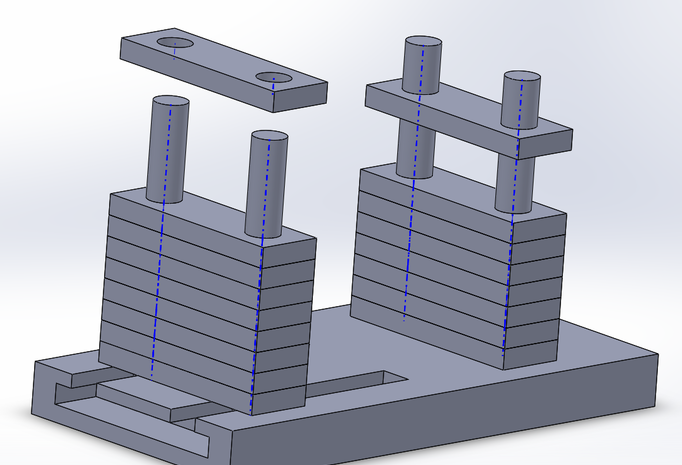
Seeing how the calculations resulted in the diagnostics being above absolute certainty, there is still a concern for how truly reliable PCR can be used to predict the development of the disease since such calculations are practically impossible in reality. However, Bayes values still must be taken at face value. Thus as of right now, using PCR to predict the development of the disease is completely reliable. The biggest source of error may have been from the small sample size given for this analysis. This sample size gave very little room for error for an accurate prediction, so a larger sample size would be needed next time. Another source of error may have been from improper lack of lighting within the fluorimeter. Without complete lack of external lighting, the data record may have caused the results to become inconclusive and thus alter the data analysis. Finally, another source of error may have come from inconsistent angles of the picture taken in the fluorimeter. different camera angles may have resulted in different data recorded from imagej which may result in inaccurate conclusions. Intro to Computer-Aided Design3D Modeling Our team decided to utilize Solid Works because it is a software that we are comfortable with and familiar with using. Due to the prior use of the Computer-Aided Design on Solid Works, the creation of our 3D digital design was a fairly smooth process. We found Solid Works to allow more freedom with the design process as it was simple to design exactly what we designed. We were able to decide on the design dimensions of our phone stand in order to maximize the function of the device. Solid Works supported the design of our new device by being simple to use and easy to navigate the software.
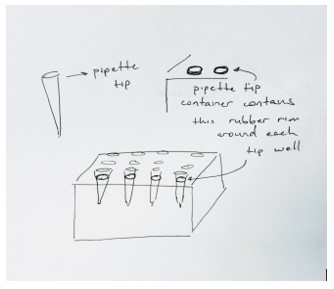
Feature 1: ConsumablesThe aspect of the experiment that we focused on improving was imaging of the fluorimeter experiment. Because we are only changing aspects regarding imaging, our consumables would remain the same and the experiment kit would include all of the standard consumables that would normally be included.This means that the tubes, glass slides, and reagents would not need to be changed due to our proposed modification. However, if we could improve anything within the consumables category, we would suggest rubber rims to be placed around all of the pipette tip wells. Our team struggled with keeping the sterile pipette tips inside their container. This resulted in an excess of hazardous waste and unused pipette tips being thrown in the trash. If rubber rims would have been placed around the wells keeping the pipette tips from easily falling out of their container, we would have been able to greatly reduce waste and save time. Feature 2: Hardware - PCR Machine & FluorimeterOur improvement focused only on imaging of the fluorimeter, so our Open PCR machine would not be affected in any way. This means that most of the materials, set-up, and procedure for the Open PCR machine and system would remain unchanged. Our one major change is the introduction of a new, more functional, phone stand to account for any sized phones. With this introduction the procedure and all other materials would remain the same. The primary purpose of the change in phone stands is to allow for high quality, easy to take pictures so students don't have to try to take a picture of the same angle while having to move the phone every time.
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||