BME100 s2018:Group4 W1030 L4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | ||||||||||||||||||||||||||||||||
OUR TEAM
LAB 4 WRITE-UPProtocolMaterials
PCR Reaction Sample List
- Denature at 95ºC for 30 seconds - the DNA helix separates to create two single-stranded molecules of DNA - Anneal at 57ºC for 30 seconds - the primers attach to the single-stranded molecules of DNA - Extend at 72ºC for 30 seconds - the DNA polymerase is activated, attaches to primers, and begins to add complementary nucleotides
Research and DevelopmentPCR - The Underlying Technology There are four main components of a PCR reaction, which are Template DNA, Primers, Taq Polymerase, and Deoxyribonucleotides (dNTPs). Template DNA is a sample of DNA used to copy multiples of it. The function of primers are to attach to sites on DNA to the DNA strands that are at either end of the segment one would want to copy. The function of Taq Polymerase is to read the DNA and attach matching nucleotides to create DNA copies. And deoxyribonucleotides (dNTPs) are the building blocks that link to form a strand of DNA. Thermal cycling is a process in which involves all of the above components. During the initial step, the DNA double helix structure unwinds. Then the DNA splits into 2 single stranded DNA molecules, which is during the denature step at 95 degrees for 30 seconds. Then, Extend at 72 degrees for 30 seconds is where the DNA polymerase binds to the primer and then adds complementary DNA nucleotides to the template strand until polymerase reaches the end and falls off. Then in the final step, the prior 3 steps are repeated to create new cycles and to make additional new DNA strands and cycles that come afterwards will strictly only have the desired DNA fragments. In the final hold, DNA rewinds back to original double helix structure. DNA is made up of four nucleotides, which are Adenine (A), Thymine (T), Cystosine (C), and Guanine (G). The first step is during Anneal because the temperature is cool enough that nucleic acids can pair with each other but 57 degrees favors the primers to bind to the template strand of DNA and add the first few base-pairs before each template strand can bind with each other again. Next is Extend at 72 degrees because that this point the temperature is more favorable for base pairing so DNA polymerase binds to the primer and then adds complementary bases to the template strand until completion.
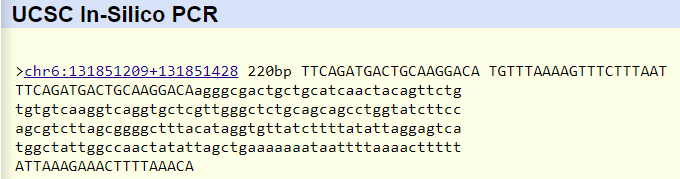
SNP Information & Primer DesignBackground: About the Disease SNP A nucleotide is a compound that is formed of a nucleoside that is binded to a phosphate group, and these nucleotides then become structural units for Deoxyribonucleic acid (DNA) [[1]]. A polymorphism is the variance of genetics within a population, where natural selection can then apply. Therefore, a single nucleotide polymorphism (SNP) is the variance of a single nucleotide or between nucleotides within the genome within a species. The given SNP is rs1044498, which is found in humans (homo sapiens), and this variation is found on chromosome 6. The listed clinical significance of this SNP is pathogenic or rather to demonstrate whether a person is benign or at a risk factor of type 2 diabetes and obesity in relation to insulin resistance[[2]]. The ENPP1 stands for ectonucleotide pyrophosphatase/phosphodiesterase1 [[3]]. The function of ENPP1 protein is to control cell response to the hormone insulin, which plays a role in the regulation of sugar levels throughout the body [[4]].Some other functions include exonuclease activity, insulin receptor binding, and protein homodimerization activity[[5]]. An allele is defined as a variant form of a gene, which arise by mutation and are found at the same place on a chromosome. The non-disease allele contains the codon AAG on the given SNP. A change is this allele, specifically at the A position, is liked to the disease. The disease-associated allele contains the codon CAG [[6]]. The numerical position of this SNP is 131851228. Primer Design and Testing Non-disease forward primer (20nt): 5' TTC AGA TGA CTG CAA GGA CA 3' Non-disease reverse primer (20nt): 5' TGT TTA AAA GTT TCT TTA AT 3'
Disease reverse primer (20nt): 5' TGT TTA AAA GTT TCT TTA AT 3' The primer test showed that the primers were located on chromosome 6 and were 220 base pairs apart, which is what was found in the primer design as they were found 200 base pairs apart and were both 20 bases long. The disease primers display no match in the results, which makes sense due to those primers not intended to be found along the human genome as it is a part of the SNP mutation.
| ||||||||||||||||||||||||||||||||