BME100 s2018:Group2 W0800 L4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | |||||||||||||||||||||||||||||||||||||
OUR TEAM
LAB 4 WRITE-UPProtocolMaterials
Research and DevelopmentPCR - The Underlying Technology
What is the function of each component of a PCR reaction? The template DNA is the original copy of DNA which is used as the variation in the experiment. Each set of template DNA should vary from organism to organism. Primers are short strands of DNA or RNA which bind to specific points on DNA. These primers are like beacons which show where replication should begin. Taq polymerase is a polymerase which operates under a specific temperature but can survive under extreme temperature variations. Deoxyribonucleotides are the individual building blocks that make up DNA.
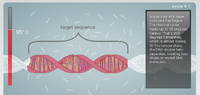
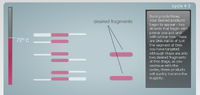
First the DNA helix is separated by increasing the temperature to 95°C. After the helix is separated the DNA is cooled to 57°C so that the primers can attach to the single stranded DNA. The temperature is then raised to 72°C in order for the DNA polymerase to attach to the primers and then begin synthesizing the correct nucleotides onto the DNA. The polymerase continues till the entire strand is covered. This process is repeated until there is an abundance of the desired DNA fragment.
In DNA, adenine pairs with thymine and cytosine pairs with guanine. During which two steps of thermal cycling does base-pairing occur? Explain your answers. When primer attaches (Temp drops to 57℃) and when DNA polymerase is working (Temp is 72° C). The primer must match with the correct nucleotides. This shows that the bases are paired according to their nucleotide sequences. DNA polymerase must match the correct nucleotides with the existing DNA which also shows that the nucleotides must match.
SNP Information & Primer DesignBackground: About the Disease SNP The rs1044498 disease SNP, present in "Homo sapiens", is connected to bone disorders that are associated with type 2 diabetes mellitus (T2DM). Only one swap in the nucleic base causes this disease SNP. The one change in the nucleic base changes the codon which is read. The codon which is associated with our disease is CAG. The numerical position of the disease SNP is 131851228.
The diseased strand had one nucleotide that was changed. We then used the genome database and entered the two different sequences to determine which one was the diseased SNP.  
| |||||||||||||||||||||||||||||||||||||