BME100 s2018:Group1 W1030 L4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | |||||||||||||||||||||||||||||||
OUR TEAM
LAB 4 WRITE-UPProtocolMaterials
PCR Reaction Sample List
Research and DevelopmentPCR - The Underlying Technology A PCR reaction has four main components: Template DNA, Primers, Taq Polymerase, and Deoxyribonucleotides (dNTP's). Template DNA provides the desired genetic sequence to be copied. The Primers attach to the desired single-stranded DNA segment at sites on either end of the desired genetic sequence. Taq Polymerase is a certain type of DNA polymerase able to withstand the high heat of PCR reactions necessary to denature DNA strands. It locates primers attached to DNA and reads the genetic code of the DNA to attach complimentary nucleotides to that strand. Lastly, the dNTP's are the components that make up the DNA's genetic code, and are the building blocks used to create copies of the desired genetic sequence. Within a PCR reaction, there are six thermal cycling steps. The procedure begins with heating the container at 95°C for 2 minutes, which brings it to the necessary temperature for the PCR process to begin. The next three - Denature, Anneal, and Extend - are the majority of the process, and repeat for a total of 30 cycles. During the Denature step, the container is heated to 95°C for 30 seconds, causing the double-stranded DNA to lose its double helix shape and separate into single-stranded DNA. In the Anneal step, the container is cooled to 57°C for 30 seconds. At this temperature, the single-stranded DNA molecules attempt to pair up, however, the large concentration of primer sequences attach to the strands before they can rejoin. The final of the three step cycle is the Extend step, during which the container is heated to 72°C for 30 seconds. The DNA polymerase is activated at this temperature, which locates primers attached to single-strand DNA segments and begins adding the complimentary nucleotides to the strand until it falls off the end. This completes the main three step cycle, which usually repeats for 30 cycles. After this, the container is held at 72°C for 2 minutes, while the DNA polymerase finishes adding complimentary nucleotides to the single-stranded DNA segments. Lastly, the container is cooled to 4°C, allowing the single-stranded DNA to cool, pair back up into double stranded DNA segments, and reform a double helix shape. DNA is made up of four types of molecules called nucleotides, of which Adenine (A) and Thymine (T) anneal to one another, and Cytosine (C) and Guanine (G) also anneal to one another. This base pairing occurs during the Anneal and Extend steps. This is because during the Anneal step, primers base pair with single-stranded DNA segments, and during the Extend step, the DNA polymerase adds complimentary nucleotides to the single-stranded DNA segments.
(The above image demonstrates the amplification of DNA through the use of Taq Polymerase.)
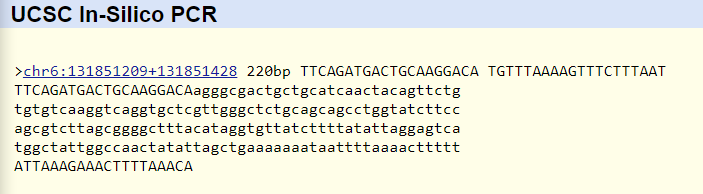
SNP Information & Primer DesignBackground: About the Disease SNP The SNP rs1044498 is found on the sixth chromosome in Homo sapiens. It is located on the gene that codes for ENPP1, or ectonucleotide pyrophosphatase/phosphodiesterase 1. The functions of this gene include 3' phosphadenosine 5' phosphosulfate binding, ATP binding, and NADH pyrophosphatase activity. The SNP occurs at the 131,851,228th position on the chromosome, where the codon AAG becomes CAG. This SNP is related to type II diabetes, and specifically bone disorders associated with type II diabetes.
Non disease forward primer: 5' TTC AGA TGA CTG CAA GGA CA 3' Non disease reverse primer: 5' TGT TTA AAA GTT TCT TTA AT 3'
Disease reverse primer: 5' TGT TTA AAA GTT TCT TTA AT 3' The primer test demonstrated that the primers were located on the correct chromosome and in the correct location. The non disease primers were located on chromosome 6, 220 base pairs apart. This is the correct distance because the forward primer and reverse primer were separated by 200 base pairs, and were 20 base pairs long. The disease primers were not recorded to exist in the human genome. This makes sense because those primers were the result of the SNP mutation. | |||||||||||||||||||||||||||||||