BME100 s2018:Group11 W0800 L4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | ||||||||||||||||||||||||||||||||||
OUR TEAM
LAB 4 WRITE-UPProtocolMaterials
As a final step, the PCR tubes will sit at 72 degrees Celsius for 2 minutes. Once the process is complete, the PCR tubes will be held at 4 degrees Celsius.
Source of Image: http://slideplayer.com/slide/273138/
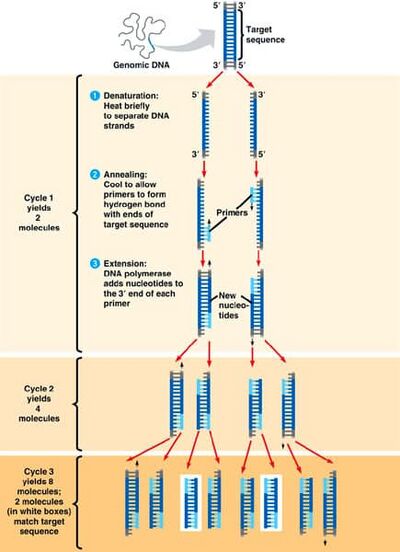
Research and DevelopmentPCR - The Underlying Technology 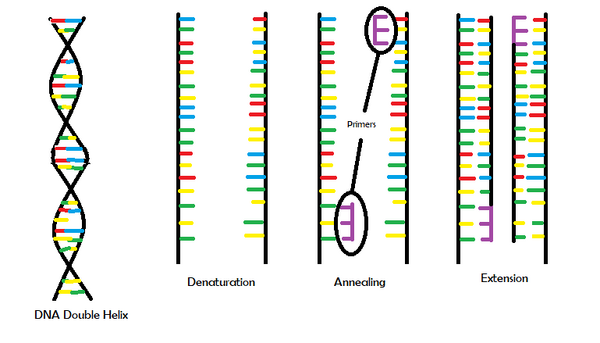 Functions of the PCR Components There are four main components involved in Polymerase Chain Reaction (PCR). One such component is template DNA. Once the DNA double helix unwinds during the reaction, one of the two DNA strands becomes the template DNA strand in which RNA nucleotides match up base pairs with (RNA nucleotides include adenine, cytosine and guanine like DNA nucleotides but instead of thymine, RNA has uracil). Primers are another part of PCR. The primers copy very specific DNA sequences by attaching to either end of the DNA that needs to be copied. Then taq polymerase plays its role. Taq polymerase is an enzyme that copies DNA and makes new strands using the existing ones. These are used for PCR because they do not break down at high temperatures. The last components involved are deoxyribonucleotides (dNTP's). These are the building blocks for the new DNA strands formed during PCR. All of these components come together to perform PCR.
There are several key steps involved in the thermal cycling process. During the first step, the samples are heated to 95 degrees Celsius and remain at this temperature for 2 minutes. During this stage, the DNA double helix unwinds. Next, the samples denature. In this phase, the temperature remians at 95 degrees Celsius but for only 30 seconds. In this process, two single-stranded DNA molecules are created from the separation of the unwound DNA double helix. The samples are then go on to the next step in the thermal cycling process, annealing. The samples are cooled to 57 degrees Celsius and remain at this temperature for 30 seconds. In the annealing process, the primers bind to the complementary sequences of the single-stranded DNA to indicate which portion of the DNA needs to be copied. The following stage is known as the extension stage. The temperature is raised to 72 degrees Celsius for 30 seconds in which the DNA taq polymerase activates and locates the primers. The temperature remains at 72 degrees Celsius for 2 more minutes as taq polymerase adds complementary nucleotides to the template DNA strand until it gets to the end of the strand and falls off. In the final step, the samples are held at 4 degrees Celsius to preserve the samples. This process is repeated at least 3 times to produce the desired fragments.
During the thermal cycling process, a crucial step known as annealing occurs. Only certain DNA nucleotides bind with others and the sequence in which they bind codes for certain characteristics. There are only four DNA nucleotides which are adenine, cytosine, guanine and thymine. Among these four, thymine bonds with adenine and cytosine bonds with guanine.
Base-pairing during thermal cycling occurs in two steps. Throughout the annealing phase, the primers base-pair complementary sequences on the single-stranded DNA molecules.The subsequent step also includes base-pairing. During the extension stage, base-pairing occurs as the temperature approaches 72 degrees Celsius. The DNA tpolymerase locates the primers to add nucleotides to the template DNA strand until it reaches the end of the strand and falls off.
SNP Information & Primer DesignBackground: About the Disease SNP DNA is composed of several smaller units called nucleotides. All nucleotides have a 5 carbon sugar and a phosphate group, but what distinguishes nucleotides from one another is their nitrogenous base. There are four different bases: adenine, thymine, cytosine, and guanine. When a single-nucleotide polymorphism (SNP) occurs, one of the nucleotides in the DNA sequence is replaced by one with a different base. The impact of an SNP depends on its position on a person's gene. The SNP rs1044498 is found is found at the 6:131851228 chromosome on the ENNPP1 (ectonucleotide pyrophosphatase/phosphodiesterase 1) gene. This gene is responsible for ATP binding, NADH pyrophosphatase activity, and calcium ion bonding among other functions. When the SNP rs1044498 occurs, the base adenine is switched for cytosine at the position previously described. The non-disease allele AAG thus becomes a disease associated allele CAG. This SNP is associated with type 2 diabetes as well as health problems related to type 2 diabetes including bone disorders.
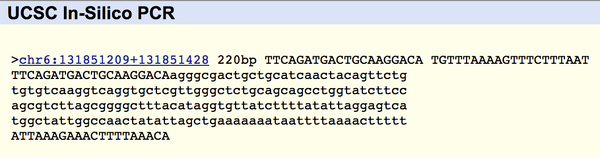
The primers tested were the non-disease forward primer 5'TTCAGATGACTGCAAGGACA and the non-disease reverse primer 5'TGTTTAAAAGTTTCTTTAAT. When tested, the result was the 220 bp sequence starting 20 bases before the SNP rs1044498 location and ending 200 bases after it, confirming that the primers we used were correct. The result window can be found below:
| ||||||||||||||||||||||||||||||||||




