BME100 s2018:Group10 W0800 L4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | |||||||||||||||||||||||||||||||||
OUR TEAM
LAB 4 WRITE-UPProtocolMaterials
OpenPCR program
Denature at 95°C for 30 seconds, Anneal at 57°C for 30 seconds Extend at 72°C for 30 seconds
Research and DevelopmentPCR - The Underlying Technology Function of PCR Reaction ComponentsThere are four (4) total components that make up the PCR reaction. First, there is the template DNA which acts as the original source of DNA for the reaction. In other words, this is what is going to be copied in the reaction. Primers, which are short pieces of DNA made in the laboratory, are working as the starting point for DNA replication. Because primers are custom, any arrangement of nucleotides can be made in the lab. Primers are necessary for the PCR reaction because DNA polymerase are unable to attach onto any old piece of the DNA, they have to attach to existing DNA in order to replicate. DNA polymerase, as previously mentioned, replicate the DNA. Nucleotides are the building blocks of what the DNA is made up of. DNA polymerase grab onto nucleotides floating around and add them to the primer. The added onto parts of the template DNA are known as deoxyribonucleotides or dNTPs. Components During Thermal CyclingThe initial step of the lab heats up the DNA for denaturation at 95°C for 2 minutes. On denaturation is reached, hydrogen bonds are broken between bases and separate strands for 30 seconds. Following this, the primers bond to 3’ end of DNA as they anneal at 57°C for a total of 30 seconds. Next, the DNA is extended at 72°C for 30 seconds in which the taq polymerase binds and copies the DNA. This brings upon the final step where the DNA continues to copy at 72°C for 2 minutes until the desired amount is reached. Finally, there is the final hold at 4°C which allows for short-term storage of PCR DNA. Base-PairingDNA is made up of four types of molecules, known as nucleotides, given the shortened names of A, T, C, and G. A being for adenine, T for thymine, C for cytosine, and G for guanine. The base-pairing of these molecules, allowing the pairs to stick together, is driven by hydrogen bonding. The pairing of molecules follows the pattern of A pairing with T and vice versa, and C pairing with G and vice versa. This pairing of molecules makes up the nucleotides, which makes up the DNA. Two Steps in Which Base-Pairing OccursThe first step that includes base pairing is extending/elongation because this is the step that involves the Taq polymerase binding to the primers on the DNA template and will begin to add the base pairs to the template DNA until the cycle ends.The other step that involves base pairing is the optional final hold step,even though this step is completely optional, it allows for any remaining base pairs to bind to the remaining DNA template in the mixture.
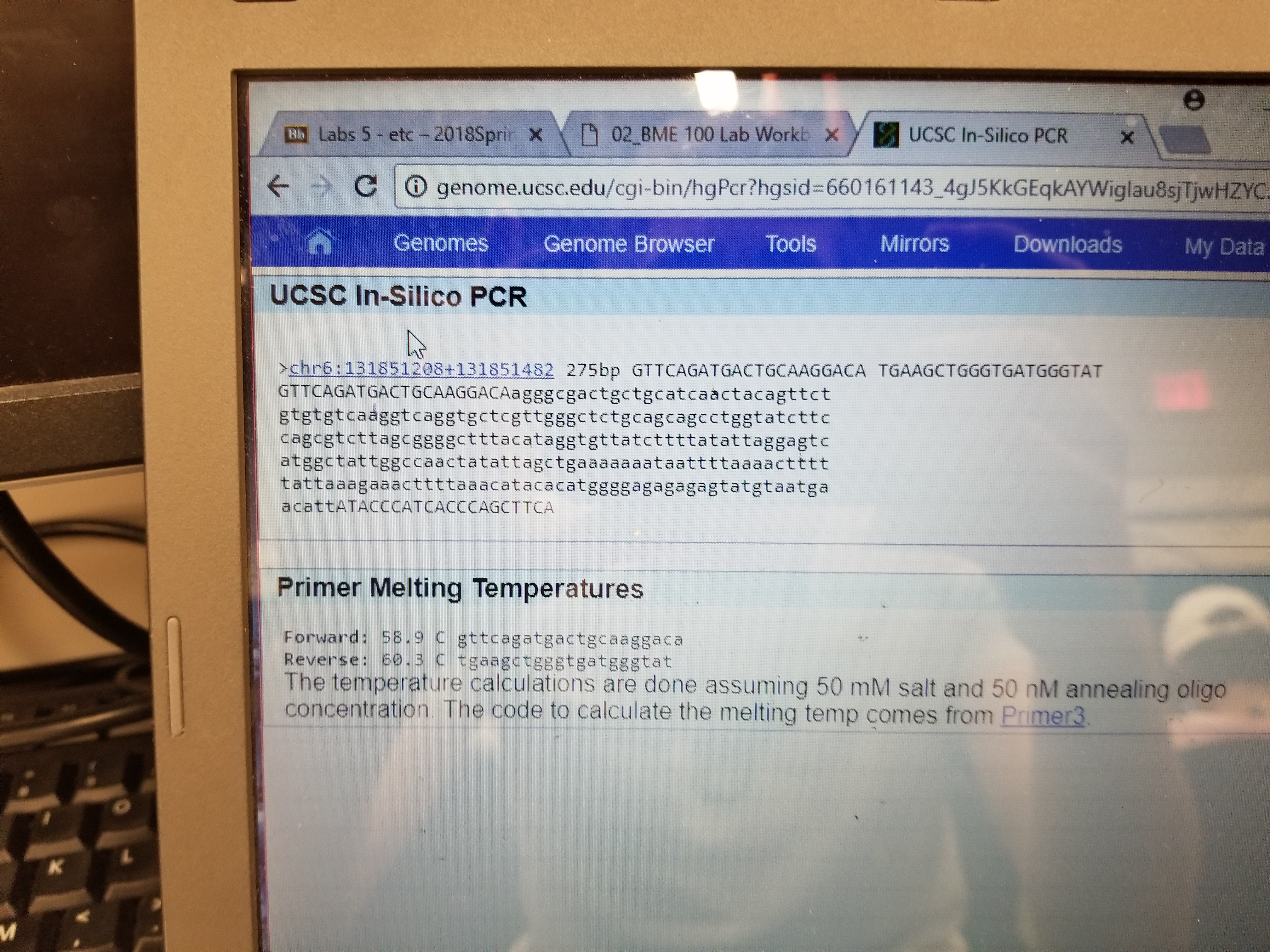
SNP Information & Primer DesignBackground: About the Disease SNP SNP, or Single nucleotide polymorphisms, are common variations in the sequence of DNA (in the nucleotides) among individuals. Sequence variations exist at defined positions associated with the individual's phenotype characteristics. Physical mapping is used to find the location of the variation and this information is then used to complete a functional analysis to study the implications of the variation. The variation researched in this lab, rs1044498, is found within homo sapiens in chromosome 6 of the DNA. The clinical significance of this is unknown, but has been found to be associated with bone disorders and diabetes. Primer Design and Testing After completing the primer test, the result showed that the non-disease primers produced about 275 base pairs.On the other hand the diseases primers showed no results due to these primers not being in the natural human genome.
| |||||||||||||||||||||||||||||||||