BISC219/F13: RNAi Lab 8
Lab 8: Heat Shock Proteins (HSP's) and Heat Shock Factor -1 (HSF-1)
Background materials for HSP's, HSF-1 and the reporter assay are provided by Cassandra Theusch and Peter Kuhn; Elisabeth A. Cox and Theresa M. Grana.
Background
Although “heat shock proteins” are best characterized in associated with temperature, mounting evidence suggests that heat shock proteins are expressed and play vital roles during many types of cell stress. Heat shock proteins work by helping other proteins fold properly and have been classified in a functional category of proteins called molecular chaperones. Molecular chaperones catalyze the folding of a newly-translated peptide into the secondary and tertiary structures that characterize the mature protein product. Improperly-folded proteins are unstable, prone to aggregation, and dysfunctional.
Correct protein folding is extremely important for protein and cell function. . Alterations in protein structure, including secondary and tertiary structure, lead to altered protein function. Similarly, improperly or incompletely folded proteins are likely to be dysfunctional and lead to complications for the cell. It should come as no surprise that improper protein folding in cells of the human body can lead to severe diseases. For example, transmissible spongiform encephalopathies (TSE’s), such as mad cow disease, chronic wasting disease, and Creutzfield-Jakob disease, are the result of improperly-folded proteins called prions. A buildup of prions interferes with cell functions and leads to apoptosis (programmed cell death). Other protein folding diseases exhibit similar pathological mechanisms. Alzheimer’s disease is thought to result from the aggregation of improperly-folded amyloid protein. Amyloid plaques accumulate in the brain and damage surrounding neurons as they grow. Certain types of cancer are associated with molecular chaperone dysfunction, which allows tumor cells to grow abnormally and survive adverse conditions.
Heat shock factor 1 (HSF-1) is a transcription factor which is required for transcription/expression of genes encoding many subtypes of heat shock proteins. Under normal cell conditions, HSF-1 expressed in the cytoplasm migrates occasionally, in small amounts, to the nucleus to activate transcription of genes encoding heat shock proteins to carry out normal cell functions. During a cell’s response to stressful conditions, HSF-1 is activated in large amounts, polymerizes, and migrates from the cytoplasm to the nucleus to search out recognized promoter sequences. This high level expression of heat shock proteins allows for refolding of proteins following stress and recovery of protein function. The specific mechanisms and molecular triggers of HSF-1 activation and migration are still unclear. Some evidence points to certain subtypes of heat shock proteins binding with and phosphorylating HSF-1 during the stress response. Yet other HSP’s may have roles inhibiting HSF-1, although scientists are still working to identify all the facets of HSF-1 regulation. More broadly, the types of stressful cellular stimuli which activate HSF-1 also have yet to be elucidated. This is a hot area of research currently, where scientists are hoping to answer some important questions about how cells respond to stressful stimuli. Perhaps your investigations will contribute to this research!
GFP and the reporter assay
A reporter assay is commonly used to assess the activity of transcription factors within a cell. You will use reporter assays to determine functional expression of the transcription factor HSF-1. A reporter assay exploits the expression of a measurable (or visible) protein, in this case GFP. The gene encoding GFP is transgenically engineered to lie downstream of a promoter to which the transcription factor binds and initiates transcription. This engineered “transgene” can be incorporated into the genome of an organism, such as C. elegans. Therefore, when conditions are right and the transcription factor’s expression is high enough, reporter gene is transcribed, and this can be measured by the amount of reporter protein produced.
Green fluorescent protein (GFP) is a commonly-used reporter protein, and, as its name suggests, is a fluorescent protein used in countless ways to visualize and quantify cellular molecules and processes. Originally, GFP was identified in and isolated from the marine jellyfish Aequorea victoria. However, modern advances in molecular biology have produced transgenic forms of GFP to more aptly suit certain experimental requirements. Due to the explosion of discoveries through experiments using GFP, the 2008 Nobel Prize in Chemistry was awarded to Martin Chalfie, Osamu Shimomura, and Roger Tsien for the initial discovery of GFP.
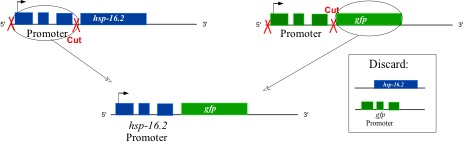
Your experiments will use a transgenic C. elegans strain which has been engineered to contain within its genome a reporter gene. This reporter construct encodes GFP under the control of a hsp-16.2 promoter. (Normally, expression of hsp-16.2 genes results in HSP-16.2 protein production.) HSP-16.2 is one subtype in the family of heat shock proteins, and therefore, it carries out molecular chaperone duties when it is present in the cell. HSF-1 binds to and initiates transcription via the promoter to hsp-16.2. Genetic engineers merely chopped out the promoter to hsp-16.2, pasted it onto the gene encoding GFP, and inserted the combo into a worm’s genome – a reporter gene was born!
Figure 1: Construction of the GFP reporter transgene. The promoter region of hsp-16.2 was pasted onto the gene encoding GFP. Therefore, transcriptional activity at the hsp promoter can be measured by the levels of GFP in the cell.
Under normal conditions, HSF-1 is present in cells, but its low level of activation cannot be directly measured using the techniques you will use in lab. When a cell is stressed, HSF-1 is phosphorylated and activated in the large amounts measurable using this GFP reporter assay. Activated HSF-1 up-regulates the transcription of the hsp16.2/gfp transgene, which allows measurement of activation through detection of GFP using fluorescence microscopy.
Lab 8: Series 3- Creating the feeding strain of bacteria for RNAi in C. elegans
In order for RNAi to be successful we must insert the plasmid expressing dsRNA of our C. elegans gene of interest into the bacteria that will make lots of double stranded RNA. We will then feed the bacteria that is full of double stranded RNA specific to our gene of interest to our worms. The process of inserting the plasmid into bacteria is called transformation. Transformation, either chemically or electrically, opens up pores or channels in the bacterial membrane that allows for uptake of the plasmid DNA.
Precise pipetting is required for transformation to be effective. Your instructor will demonstrate the correct technique. If you have any questions, please consult with her/him before proceeding with any micropipetting.
To recap what has been done in order for your to be ready to do today's work:
Yesterday you inoculated a few milliliters of LB broth containing E. coli that we are sure have maintained our genetically engineered plasmid, pPD129.36, modified to contain an antibiotic resistance gene to ampicillin and all or part of the C. elegans hsf-1 gene that you want to investigate. You added ampicillin to the broth to ensure that the plasmid DNA would be maintained by the cells. Overnight the bacteria have grown to high density and the plasmid DNA has undergone many replications. However since you started with a single colony of bacteria and that colony grew from a single transformed cell, all the copies of the plasmid DNA in your overnight culture should be identical (“clones” of one another). To isolate the plasmid DNA from this bacterial strain that expresses C. elegans hsf-1 , you will perform what is commonly called a “mini-prep”. This term distinguishes the procedure from a “maxi-” or “large scale-prep” which involves a larger volume of cells and additional steps of purification. The overall goal of each “prep” is the same--to separate the plasmid DNA from the chromosomal DNA so that a certain gene on the plasmid DNA can be studied further.
Synopsis of Today's Work
Today you will transform the isolated plasmid DNA, pPD129.36+hsf-1, into an IPTG inducible strain of E. coli called HT115(DE3), and spread the transformed bacteria onto LB agar media supplemented with ampicillin (resistance conferred by a gene expressed from the plasmid).
Part 1: Plasmid DNA Isolation
What's Happening in Your MiniPrep
To isolate plasmid DNA from an overnight culture of cells, the media is removed from the cells by centrifugation. The cells are resuspended in “Solution I” which contains Tris to buffer the cells and EDTA to bind divalent cations in the lipid bilayer, thereby weakening the cell envelope. Upon the addition of “Solution II,” the chromosomal DNA and the plasmid DNA are denatured by the sodium hydroxide, and the cellular proteins and lipids are dissolved by the detergent, sodium dodecyl sulfate (SDS). The pH of the solution is returned to neutral by the potassium acetate in “Solution III.” At neutral pH the SDS precipitates from solution, carrying with it the dissolved proteins and lipids. In addition, the DNA strands renature at neutral pH. The chromosomal DNA, which is much longer than the plasmid DNA, renatures as a tangle that gets trapped in the SDS precipitate. The plasmid DNA renatures normally and stays in a water based solution. In this way the plasmid DNA is separated from the chromosomal DNA, the proteins and the lipids of the cell. Plasmid DNA can be precipitated out of solution in absolute ethanol and then put back into solution (in an appropriate concentration) in water. The ingredients and concentrations of a stock solution of all reagents (such as Solutions I, II, and III) can be found in the Media Recipes section of this section of the wiki.
Protocols
Plasmid DNA Isolation
Only one of your two overnight cultures should appear cloudy with bacterial growth. If your control is cloudy, please inform your instructor. (You can find the ingredients and concentrations for ingredients of Solutions I, II, and III in the Series 3 Media Recipes page.
- To start the plasmid DNA isolation, label 2 microfuge tubes with pPD129.36+hsf-1 (our plasmid name + our C. elegans gene), plus your initials and team color.
- Check to be sure that your cells have not settled. If they have, mix the cultures but do NOT invert them because they will leak. Then pour some of the overnight culture so that both microfuge tubes are almost full. If you are nervous about pouring the bacteria, you can pipet 750 microliters into each tube twice, so that there is a total of 1.5 ml in each tube. The exact volume doesn’t matter but the tubes should be quite full when you close the cap.
- Place the tubes in the room temperature microfuge so that the hinges of each cap is facing out. Paying attention to this small detail will help you know where in the tube to find your pellets. While it is not essential to do this in this step, it is a good habit to get into since sometimes pellets are very small and hard to see. Be sure your tubes are balanced, then spin the tubes for two minutes at 8,000 rpm. Check the rcf speed of the centrifuge while it is spinning (by hitting the toggle that changes the readout) and record the rcf’s in your lab notebook. Relative centrifugal force (rcf) is what should be used in M&M to describe your speed since it is universal and rpm is rotor and centrifuge dependent.
- If the supernatant is not clear, recentrifuge until it is. When the supernatant is clear, pour it into the waste beaker that is on your bench, then flick the tube with the cap open to remove the last few drops of liquid. The cell pellet will not fall out.
- Resuspend your cell pellets COMPLETELY in 100 microliters of solution I. Pipet up and down until ALL of the cells are uniformly suspended in Solution I. Be sure to change tips between samples. Leave the cells at room temperature as you prepare solution II.
- To make solution II, mix 500 microliters of 2% SDS with 500 microliters of 0.4M NaOH in a microfuge tube. Close the cap and invert the tube several times to mix the contents. Add 200 microliters of solution II to each miniprep and invert the tubes five or six times to mix. In some cases the minipreps may appear to "clear" but don't worry if you don't see a big change in yours. Place the tubes on ice for five minutes.
- Add 150 microliters of solution III to each miniprep and immediately vortex each tube for 10 seconds with your vortex set at the highest setting. White clumps should appear in the solution after you vortex it. Place the tubes in the room temperature microfuge and spin for 3 minutes at the HIGHEST speed.
- While the minipreps are spinning, label another set of microfuge tubes with the name of the plasmid (pPD12936+hsf-1)and your initials or team color. The new tubes can be left at room temperature.
- A white pellet should be visible when you remove your minipreps from the microfuge. Use your P1000 to transfer 400 microliters of each supernatant to the appropriate clean microfuge tube. It's OK to leave some of the supernatant behind. Try to avoid transferring the white pellet.
- Add 1 ml of room temperature 100% ethanol to each new tube. The tubes will be quite full. Close the caps and invert the tubes at least five times to thoroughly mix the contents.
- Spin the tubes in the room temperature microfuge for 2 minutes at HIGHEST speed. It is important to orient your tubes so that the hinges are up this time, as the pellets are expected to be barely visible.
- Pour off the supernatants into the waste beaker on your bench then flick out the last few drops. The tubes do not have to be completely dry.
- Carefully, wash the pellets with 500 microliters of 80% ethanol. (You may need to make a few mls of 80% ETOH from the 100% ethanol stock provided). When you wash the pellets, add the 80% ethanol slowly and carefully, so that the liquid flows down the side of the tube away from the pellet. After the ethanol has been dispensed, immediately remove it with the same tip, making sure to keep the tip on the side of the tube that doesn't have your pellet. Some liquid will remain in the tube, and it can and must be removed using your P200, set to 100 microliters. Try to remove as much of the ethanol as you can (without removing your pellet!). The pelleted DNA will have to be completely dry before you add water or your DNA will not go into solution.
- After all the DNA pellets have been washed, put your tubes with their caps open in the SpeedVac that is at the back of the lab. If you were able to remove most of the ethanol with the P200, then after two or three minutes the last drops of ethanol will have evaporated and the pellet will be barely visible. If the DNA pellet is invisible please see your instructor to determine how much water to use in the next step.
- If you have a visible pellet of DNA, add 30 microliters of sterile water to each tube and vortex each tube enough for all of the DNA to be off the wall of the tube and into solution. The pelleted plasmid DNA must dissolve completely in the water. The liquid can be brought back to the bottom of the tubes by spinning for a few seconds in the microfuge. You can then combine the dissolved plasmid DNA into one of the two tubes so that you have approximately 60 microliters of plasmid DNA as one aliquot. Store the DNA on ice until you are ready to use part of it and then give the rest to your instructor to freeze. DO NOT DISCARD ANY OF YOUR PLASMID ISOLATE!
Part 2: Measuring the concentration of plasmid DNA using the NanoDropper
Proper transformation requires the concentration of bacteria to plasmid DNA to be right. We will use a nano-dropper to determine the concentration of the plasmid DNA from a previous plasmid isolation procedure.
Use the ThermoScientific NanoDrop Spectrophotometer in L308 to measure DNA by taking Absorbance at A260nm. This spectrophotometer uses only 1 microliter of sample and does not require cuvettes. The sample is held in place by fiber optic technology and surface tension that holds the sample in place between two optical surfaces that define the pathlength vertically and dynamically. Measurement can be assessed in a range of 2 to 3700nm/microliter dsDNA. These are expensive machines so make sure you follow the directions carefully and ask your instructor for guidance as needed.
More information is available from the manufacturer's website at: | http://www.nanodrop.com/HowItWorks.aspx
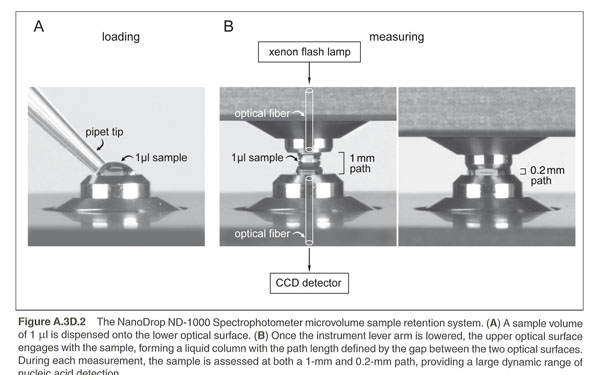
Using the Nanodropper
1. Clean the upper and lower optical surfaces of the sample retension device by pipetting 2 microliters of clean deionized water onto the lower optical surface. Close the lever arm and tap it a few times to bathe the upper optical surface. Lift the lever arm and wipe off both optical surfaces with a Kimwipe.

2. Open the NanoDrop software from the Desktop of the computer and select the nucleic acids module.
3. Initialize the machine by placing 1 microliter of clean deionized water onto the lower optic surface, lower the lever arm, and select initialize from the NanoDrop software. Once initialization is complete (~10sec.), clean both optical surfaces with a Kimwipe.

4. Perform a blank measurement by loading 1 microliter of purified deionized water.
Note that as in traditional spectroscopy, the blank will be subtracted from subsequent measurements. If you want to determine the contribution of a specific buffer or diluent, measure the buffer first using distilled water as a blank. If the buffer does not contribute to the A 260nm reading, then deionized water is fine to use as the blank. The water or buffer should always be measured to be sure that the instrument has been zeroed properly. The measurement of water or buffer should be zero or very close. All measurements are automatically normalized to 340nm.
5. Measure the nucleic acid sample by loading 1microliter of sample and selecting "measure". Record your DNA concentration in your lab notebook and on the tube of plamid DNA. Once the measurement is complete. Clean both optical surfaces with a Kimwipe and the machine is ready for the next sample.
You should ensure that the appropriate constant (50 for dsDNA or 40 for RNA) has been chosen. The software automatically calculates the nucleic acid concentration. If the calculation is done by hand, the A260nm is represented as a 1cm path for convenience, even though 1-nm and 0.2nm paths are actually used during the measurement cycle.
Clean Up
When the last sample was been measured, clean the sampling device by repeating step 1.
Part 3: Transformation of plasmid DNA into E. coli strain HT115(DE3)
The competent HT115(DE3) bacterial cells are on the instructor’s bench. You will transform some of your plasmid DNA into these bacteria. The cells are very fragile, so treat them gently.
The genotype of the HT115(DE3) cells is: F-, mcrA, mcrB, IN(rrnD-rrnE)1, rnc14::Tn10(DE3 lysogen: lavUV5 promoter -T7 polymerase) (IPTG-inducible T7 polymerase) (RNAse III minus). This strain grows on LB or 2XYT plates. This strain is tetracycline resistant. The most important things about this strain is:
- It has an IPTG inducible T7 polymerase. The chemical IPTG (Isopropyl β-D-1-thiogalactopyranoside) is a lactose mimic and is commonly used to induce the production of large amounts of protein. In this case when the cells are incubated with IPTG lots of the polymerase T7 is made. This T7 polymerase binds to T7 specific promoters on the plasmid and transcribes lots of our gene of interest.
- RNAse III minus. The strain is lacking the RNAse III enzyme which typically breaks down double stranded RNAs. We want double stranded RNAs to accumulate.
The prep staff prepared these competent HT115(DE3) cells for you using the Inoue Method Media:Inoue bacterial transformation.doc. Reference: Inoue H., Nojima H., and Okayama H. 1990. High efficiency transformation of Escherichia coli with plasmids. Gene 96: 23-28. The cells were made competent to take up free plasmid DNA by this treatment. This treatment makes the cell wall and membrane more permeable and our transformation efficiency much greater but it weakens the cells; therefore you must keep them cold and mix them gently throughout the transformation (no vortexing!).
- Obtain a tube containing 50 μL of competent cells from your instructor.
- Label the top or the side of the tube with HT115(DE3), pPD12936+hsf-1, and your initials or team color.
- Consult with your instructor to determine how much of your plasmid DNA you should add to the cells to start the transformation. When you have determined the correct vol., pipet between 1μL and 10μL of your plasmid DNA to the tube. (The volume you should use is dependent on the conc. you achieved in your mini-prep.) Pipet up and down once to mix the DNA and the cells. Close the cap and let the transformation mixture sit on ice for 10 minutes.
- Heat shock by incubating the transformation mix at 42°C for 90 seconds, exactly. This step must be timed exactly. Remove the tube at the end of 90 seconds directly into to your ice bucket (carry your ice bucket over to the heat block) while you get your LB ready.
- Consult your instructor to determine the volume of warm LB broth you should add (usually between 250-500 microliters) to the transformation mix. LB aliquots can be found in the 37°C incubator near the lab door. When pipetting the media, remember to release your thumb on your micropipet slowly, to avoid splashing the liquid on the end of the barrel. The barrel is not sterile and if you see the liquid touch it, then discard the media in the waste beaker and try again with a new tip.
- Once you have added the LB, close the cap and invert the tube once or twice to mix the contents. Incubate at 37°C for 30-60 minutes in the bench top incubator/ mixer with occasional gentle mixing.
- While the plasmid DNA is being taken up by the competent cells and the new genes provided by the plasmid are being expressed by the bacteria, label one LB + amp agar plate and one LB + amp + tet plate. Label the bottom of the two plates with the strain's identity (HT115(DE3)), the plasmid used (pPD12936+hsf-1), the date, your initials and team color. You must label the bottom of the plated since the tops are easily switched. Put these plates in the hood with the blower on and with the lid ajar to dry the surface of the agar for about 10 minutes or until the surface looks dry but is not badly dehydrated.
- Once the transformation mix has incubated at 37°C for 30-60 minutes, invert it to mix the contents and pipet 100 microliters of transformed cells onto the center of each of your labeled plates. Pour 5-10 glass beads onto the plates. Put the lid back on and gently swirl the beads all over the agar to spread the transformed bacteria around. When you are done pour the beads into the beaker with disinfectant near the sink.
- Replace the lids and leave the agar plates undisturbed for a few minutes to allow for the liquid to be absorbed by the agar.
- Once they have dried enough that the surface does not appear wet, invert the plates (lid side down) and incubate them at 37°C for 24 hours. The plates should be incubated with the agar side up so that condensation will not drip onto the surface of the agar and smear the colonies that will be growing there.
- Save the remaining transformation mix for 24 hours or until we are sure that there is at least one colony growing on each of your plates.
What would it mean if you had no colonies on your plate? Normally, you would expect to have around 100 pale color colonies on each plate. If you have at least one well isolated colony on the plate, you’re all set. After the 24 hour growth period the plate should be placed in the rack in the refrigerator labeled with your lab day. You will use a single colony from the plate to make an overnight broth culture on the day before next lab. If you have no colonies on one or more of your plates, please notify your instructor right away.
Before leaving lab today, give the rest of your isolated plasmid DNA to your instructor in a labeled microfuge tube. Make sure your tube is labeled with your name, lab day, plasmid name, DNA concentration and color coded with a piece of tape in your team color.
To do on the day before the next lab
You and your partner will return to the lab to make an overnight broth culture of your selected colony as described below. This process will create a sub-culture of many identical copies of the bacteria containing the plasmid carrying the construct to RNAi the gene that you want to study.
- Find your plate in the glass front refrigerator in a rack labeled with your lab day. Select a colony to start your overnight culture. At this point, ALL of your colonies should contain your plasmid of interest.
- Begin by obtaining two tubes of LB broth (each will have 5 ml of broth) from the refrigerator in the back left hand corner of the room.
- Add 5 microliters of the 50mg/ml ampicillin stock (also found in the refrigerator with the broth) to each tube. Calculate the effective concentration of ampicillin that you have in your LB tube (remember V1 x C1= V2 x C2) and record that information in your lab notebook.
- Gently swirl your LB +amp broth to mix the contents.
- Label the two sterile glass culture tubes with tape in your team color. Label one with "pPD129.36 hsf-1" and your initials. Label the other with your initials only.
- Inoculate the broth with your bacteria by using a sterile disposable loop to scrape your candidate colony off the plate. Be sure not to touch the plate with the loop except on the desired colony and don’t pick up any satellite colonies! Gently swirl the loop in the LB+amp broth - you should be able to see the colony come off the loop. The second tube of broth labeled with just your initials is a control and should not be inoculated with bacteria as it is your control for contamination.
- Balance the 2 tubes across from each other on the rotating wheel in the 37C incubator at the front of the room when you come in the door.
- Incubate these broth cultures at 37°C overnight. Do not forget to make sure the wheel is rotating when you leave!