BISC209/S13: Lab5
LAB 5: Soil Community Bacterial Member Identification by 16S rRNA gene sequencing
Hopefully, this week, you have pure cultures from your enrichment/selection and isolation work. Make sure that these potentially pure cultures have colonies that look like the original source colony and that all of the colonies look the same (they can be of different size). If you are having trouble obtaining a pure culture, consult with your instructor BEFORE LAB 5. We can't identify your isolate by 16S rRNA gene sequencing if it is not a pure culture.
Background
Today you will begin the identification of culturable bacterial isolates in your soil habitat. Using your culture isolates as the template, you will amplify, in a polymerase chain reaction (PCR), part of the 16S rRNA gene from your pure cultures of bacteria using "universal" bacterial primers and a proof-reading DNA polymerase. This gene is an excellent phylogenetic marker for bacteria and it has been widely used in culture-independet surveys of microbial community composition and in bacterial identification. If you are unfamiliar with the basic theory behind pcr amplification of target DNA, please review it in at the following web site: PCR animation http://www.dnalc.org/resources/animations/pcr.html .
We will use the bacterial primers that target conserved part of the 16S rRNA gene, called 27 Forward (27F) and 1492 Reverse (1492R). The numbers refer to positions on the Escherichia coli 16S rRNA gene. The direction of elongation of new DNA can only proceed in the 5' to 3' direction; therefore, the primer that anneals to base 27 on the 16S rRNA gene template is called the forward primer because DNA synthesis moves downstream on the template. The 1492R primer anneals to base 1492 on the opposite template strand and DNA synthesis moves upstream from the template. Our primers are called "universal" bacterial primers because they anneal to a 16S rRNA region that is highly similar (conserved) to most Bacteria. If we were interested in identifying eukaryotic microbes, such as fungi or protozoa, or if we wanted to identify the Archaea (which are neither Eukarya or Bacteria), we would have to use different primers.
Although our primers have sequences of single stranded DNA that are common to the 16S rRNA gene of most bacteria, the section of the 16S rRNA gene that will be amplified by the DNA polymerase contains highly variable regions of that gene. So variable, in fact, that we should be able to use the DNA sequence of those ~1,500 bases to identify our soil bacteria by comparing the sequences we amplify to the same region of the gene in other bacteria. 16S rRNA gene sequences are archived in an extensive, public database called the Ribosomal DNA Project (RDP) found at | http://rdp.cme.msu.edu/index.jsp;jsessionid=04A058D492CD00120AC01A70AAD7624A . There are so many 16S rRNA gene sequences archived at this site that we should be able to use their powerful, public search engine to identify our soil bacteria down to the genus or species level, if our pcr and the sequencing of the gene from the clones all works well.
Part A: Prepare Lysates from Pure Cultures of Bacteria of Interest
Your careful attention to following the directions exactly in each of the procedures involved in this process will go far to insure the project's success. Keep in mind that the reagents involved are expensive and limited. The best way to end up with good data is to read ahead, think about what's happening in each reaction step or in each part of the process, create carefully prepared and organized flow diagrams or outlines in your lab notebook BEFORE you come to lab, ask questions BEFORE you commit yourself if you are unsure about a step, and focus on the task at hand.
PROTOCOL:
1. Each student will obtain DNA from 4-7 unique bacteria (about 16 per team). Use well isolated colonies from your best plate (one that's unquestionably a pure culture) for each of your bacterium of interest. Consult with your partners to make sure that your team is identifying bacteria that are unlikely to be of the same species and NOT likely to a nonbacterial microbe (e.g. mold).
2. Each students needs PCR tubes of your team color (0.2 ml vol.). You will need one for each of your isolates. Wear new gloves when you take them out of the jar on the instructor bench and label each with a unique code to indicate each isolate you want to id. Be incredibly careful and DO NOT CONTAMINATE THE INSIDE OF THE CAP with your skin flora. Use your sampling habitat id (site A, B, C, or D) and a unique set of numbers that your lab instructor will assign each student. Numbers will be found on the board.
3. Using your P200 micropipet, pipet 20 μL of sterile water into each of the prelabeled PCR tubes.
4. Touch a well isolated colony from a pure culture with a sterile P10 micropipet tip (the tiny ones, not the P20 tips-use the tip as if it were a toothpick) and resuspend a barely visible amount of bacteria adhering to the tip by swishing the tip around in the appropriately labeled tube with water. Resist the urge to pick up too much cell material, but be aware that some of your isolates, particularly those dry, powdery Actinomycetes (like Steptomyces) are hard to get any cells to adhere. For those, it is ok to take a bit more. The tiniest bit is enough, but make sure there is some part of the colony going into the lysate. Putting in too many cells can inhibit the PCR reaction and be as bad as putting in none.
5. Repeat for your other isolates-- into separate tubes.
6. Vortex, centrifuge, and freeze all samples at -80 ˚C for 10 minutes.
7. Boil all samples for 10 minutes at 95 ˚C in the thermal cycler. This will lyse the cells and inactivate bacterial enzymes. Be careful when you remove the tubes from the thermal cycler. Point them away from you and ease the lids open while still covering the outside of the tops with your gloved fingers. Do this slowly and carefully with the opening pointed away from you. You don't want the caps to pop and make an aerosol of your bacteria and you don't want to lose your lysate.
Part B: PCR AMPLIFICATION of 16S rRNA gene from lysates prepared above
Note: We are using a kit from New England Biolabs called Phusion® High Fidelity PCR Master Mix with HF Buffer | http://www.neb.com/nebecomm/products/productM0531.asp. Because you are using a kit to amplify the bacterial 16S rRNA gene and the manufacturer does not have to disclose how to make each reagent from scratch, you can not include typical instructions for making stock reagents (ingredients and concentrations)in your Materials & Methods section. However, since the manufacturer's website includes instructions for how to use this product, you can refer your reader to that information and explain simply that you used this kit to accomplish the gene amplification according to manufacturer instructions. Keep in mind that you WILL have to give specific information about the gene template, the primers (DNA sequences), and the thermal cycler program since those things are not universal but specific to the primers and DNA we want to copy. (Size and GC content are crucial variables). "Universal" Bacterial Primer sequences for the 16S rRNA gene that we will be using are 1492R reverse primer TACGG(C/T)TACCTTGTTACGACTT and the forward primer 27F is AGAGTTTGATC(C/A)TGGCTCAG. Note that the () indicates degeneracy, where either of two bases can be used. Our primers are purchased from Integrated DNA Technologies (IDT) at | http://www.idtdna.com/site.
All reagents for the PCR should be kept on ice and the master mix should be thawed on ice. Since DNA polymerase can function at room temp, we don't want the reaction to start until all the tubes are in the thermal cycler.
The components below have been aliquoted and prepared for you and are in PCR tubes of your team color. Label a PCR tube for each of your lysates with a fine point Sharpie on the top and side of the tube with the unique identifier for each bacterial isolate. We will set up one tube per lab as a neg control.
Setting up the PCR Mix
WEAR GLOVES AT ALL TIMES AND DON'T TOUCH THE INSIDE OF THE TUBE CAPS OR YOUR PIPET TIPS--Always use a new tip when going into anything in a PCR reaction. (Contamination is a significant problem in PCR)
Using a P2 or P10 and filter tips (remember that the P2 has two red decimal place volume indicators while the P10 only has 1 red decimal place indicator. MAKE SURE YOU HAVE DIALED IN THE CORRECT VOLUME!), add 2 µL of your frozen/boiled lysate (containing the template DNA) to the pre-aliquoted 48 µL of Master Mix (contains DNA polymerase, dNPTs, MgCl2, and buffers), primers and nuclease free water mixture described above (for a total volume of 50 μL) in clearly labeled PCR tubes of your team color. Make sure you label on both the top and sides of the tube. The tubes are tiny (holds only 0.2 ml vol) and it is hard to write on them legibly but doing so is very important. You will have to use a unique identification code for your isolates and keep the key to the code in your lab notebook (also give a copy to your instructor).
For the negative control, one person in the lab will add 20 µL of water rather than 18 µL (in place of the template DNA).
For the positive control, one person in the lab will add 19 µL of water and 1 µL of E. coli DNA extract. When you have mixed your DNA or water into the PCR mix by tapping VERY LIGHTLY or flicking to be sure that all reagents are mixed and not adhering to the tube wall, spin briefly, and take your tubes to the thermal cycler when your instructor says it's ready. Keep them on ice until then, but wipe off the bottom of the tubes before putting them into the machine. Make a template key in your lab notebook as to where in the thermal cycler you put your tubes.
Component TABLE
| Component | amt. in a 50 μl reaction |
Final Conc. |
|---|---|---|
| Purified DNAase free Water |
18 μl | |
| 2x Phusion Master Mix | 25 μl | 1x |
| 27F primer | 2.5 μl | 0.5 μM |
| 1492R primer | 2.5 μl | 0.5 μM |
| template DNA | 2 μl | 50-250 ng optimum is 100 ng of DNA/reaction |
PRIMERS:
- 27F AGA GTT TGA TC(C/A) TGG CTC AG Universal bacterial primer Lane et al. 1991
- 1492R (s) ACG G(C/T)T ACC TTG TTA CGA CTT Universal bacterial primer Lane et al. 1991
Hold the tubes on ice until your instructor tells you the thermal cycler is ready to be loaded. Wipe the outside of the tubes to remove all ice and water before placing them in the thermal cycler.
The thermal cycler program is, generally, similar for all PCR reactions, but the annealing temperature is dependent on the primer pair. When you design primers, the primer annealing temp. can be calculated based on the GC content and other factors. For 27F and 1492R, a range of 48-55 ˚C is ok, although higher temp. may lead to increased specificity that excludes some organisms' DNA from being amplified.
The length of the fragment you are amplifying determines the extension time. A general rule of thumb is to use an extension time of 1 kb per minute. Here, we amplify with primers designed for the 27th and 1492th positions in the 16S rRNA gene region based on E. coli 16S rRNA gene. Therefore our fragment is expected to be about 1.5kb long, so we will use an extension time of 1.5 minutes per cycle.
Thermal Cycler Program:
3 step program
| Cycle Step | Temperature | Time | # of Cycles |
|---|---|---|---|
| Initial Denaturation | 98 ˚C | 30 sec. | 1 |
| Denaturation Annealing Extension |
98 ˚C 55 ˚C 72 ˚C |
30 sec 30 sec 90 sec |
35 |
| Final Extension | 72 ˚C 4 ˚C |
10 min Hold |
1 |
The PCR will run for 1.5 hr or so.
Part C: Agarose Gel Electrophoresis of Clean PCR PRODUCT
To see if you successfully amplified the 16S rRNA gene and not anything else, you will "run a gel" on your PCR products. To run a gel means that we will perform an electrophoretic separation of the DNA fragments in your PCR product, using 1/10 vol. of your PCR product applied to a 1% agarose gel stained with Sybr Safe DNA stain. Your instructor will photograph the gel, label it with your amplicon id from the template and post the gel photo to the Data folder in Resources in Sakai so you can evaluate your success at 16S rRNA gene amplification. You should see a single band of ~1.5kb indicating that the only dsDNA in your PCR product came from amplification of a ~1500 bp gene fragment. Can you explain how we know the size of our amplified gene fragment?
Your agarose gel is made of 1.0% agarose solution (w/v) in 1x TGE buffer (10x=0.25 Tris, 1.9M Glycine, 13mM EDTA) with SybrSafe™ stain.
DNA is uniformly negatively charged and will, therefore, move toward the positive electrode. The separation is determined by the size or mass of the molecule or fragments of DNA.
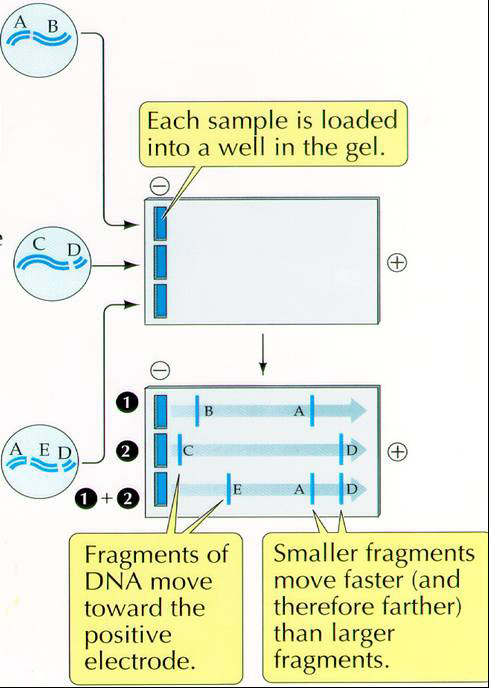
Procedure for Agarose Gel Electrophoresis of PCR products
Load 1/10 of the total volume of PCR product (1 µL minimum). In our case we should load 5 µL.
You will put the 5 µL of your PCR product as a spot on a small piece of parafilm and add a 5 µL amount of well mixed loading dye (0.25% XC, 30% glycerol, 0.1mg/ml RNAase). Mix by pipetting up and down before loading the "mix" into a lane of the 1% agarose gel (1% wt/vol in 1xTGE buffer with Sybr Safe DNA stain. Sybr Safe is a proprietary reagent from Invitrogen that cross-links a fluorescent compound to DNA and allows DNA to be visualized under UV light. It is prepared and used according to manufacturer's directions http://www.invitrogen.com). Apply your samples to the wells that you have signed up for on the gel template. Be sure to leave the first two lanes and the last lane empty for the 100 bp ladder, the positive control and the negative water control.
Loading dye contains glycerol to keep our sample in the lane rather than floating away and will have one of 3 marker dyes (bromophenol blue, xylene cyanol, or
orange G) that facilitate estimation of DNA migration distance and optimization
of agarose gel run time. 1x TGE buffer is used in this electrophoretic separation (25 mM Tris, 0.19 M glycine, 1.3 mM EDTA. The gel will be run at 120V for approximately 30 minutes.
How will you judge a successful amplification? How many fragments and of what size do you expect to see?
Make sure you give back the rest of your isolate lysate and the rest of the PCR product, and the cleaned up PCR product tube (if performed in this lab) to your instructor to freeze after the gel is loaded. Both are now in identical looking microfuge tubes with volume being the only visible difference. Make sure it is clear which is the PCR product and which is the genomic DNA isolate.
Make sure eeverything is clearly labeled with your initials, lab section (Tues or Wed), soil sample identifier code letter, your unique code information.
Your instructor will photograph and label the gel according to the template you have filled out and post the results to the data folder in Resources in Sakai.
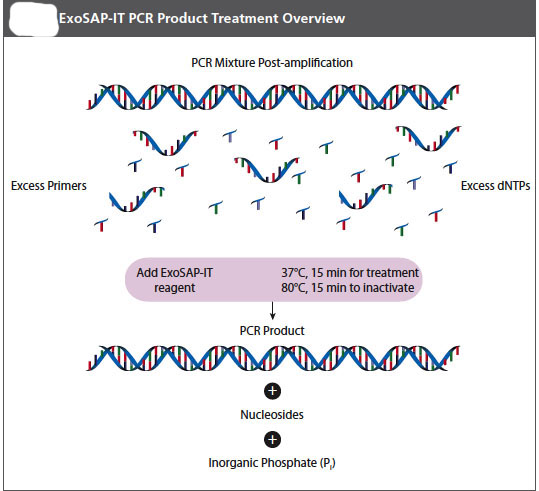
Part D: PCR Purification (Clean-Up)
It is important that your pcr product be free of left over DNA polymerase, primers, and free nucleotides before a sequencing reaction is attempted on it. Your instructor will let you know if you will do this purification today after you have loaded part of your sample on the gel or if she will do it for you later, or, perhaps, we might have the sequencing facility do it for us. The important thing for you to know is that it must be done and that there are several methods commonly used to accomplish this purification. One of the simplest is an enzymatic teatment. We will use an Exosap-it® kit from USB.
USB ExoSap-iT PCR Product Cleanup (Product Number 78200)
Protocol
ExoSap-IT reagent treats PCR products ranging in size from less than 100 bp to over 20 kb with no sample loss. The clean-up reagent is proprietary, but it is based on Exonuclease I and Shrimp Alkaline Phosphatase. Recombinant (rSAP) is added directly to the PCR product to degrade primers and to dephosphorylate dNTPs that were not consumed in the reaction. Add EXSAP_IT reagent directly to the reaction products following PCR. Since ExoSAP-IT reagent is active in commonly used PCR buffers, no buffer exhange is required.
1. Remove ExoSAP-IT reagent from -20 ˚C freezer and keep it on ice throughout this procedure.
2. Mix 8μL of a post-PCR product with 2 μL of ExoSAP-IT reagent for a combined 10 μL reaction volume.
3. Incubate at 37 ˚C for 15 minutes to degrade remaining primers and nucleotides.
4. Incubate at 80 ˚C for 15 min. to inactivate ExoSAP-IT reagent.
5. The PCR product is now purified and ready for use in DNA sequencing or other primer extension applications.
6. Treated PCR products should be stored at -20 ˚C until required.
NOTE: ExoSAP-IT reagent must be stored in a non-frost free freezer!
Part E: Submitting Amplicons for Identification
Before your next lab, your instructors will prepare a 96 well plate of your PCR products that showed successful amplification of the 16S rRNA gene. We will send these samples to a commercial DNA sequencing facility that will "clean-up" our PCR products by removing primers, polymerase and unused dNPTs that might inhibit the reaction. They will optimize the pH and the concentration of the DNA for chain termination sequencing of your amplified 16S rRNA genes. The sequencing reaction will be done for you and the resulting DNA fragments will be separated by size in an automatic sequencer with a laser that can read the fluorescent tags on the end of each fragment. This crucial part of identification of bacterial members of your soil community will be done for you and the DNA sequences will be reported back as .ab1 files that will require special software to analyze. We want you to understand exactly what's going on in those automatic sequencing machines. Make sure you understand how chain termination (Sanger) sequencing works. We should have the sequencing data back by the time we are scheduled to learn how to analyze the data so we can find out, we hope, the identity of a few members of the bacteria in your soil community. In 2013 we will use: | Genewiz's sample submission guide
Characterization of Soil Bacteria: Maintaining Pure Cultures of Isolates
Continue to subculture your pure isolates to fresh NA plates each week (isolation streak a well isolated colony onto a fresh plate), in the next lab you will make a bacterial smear and do a Gram stain and start other tests to provide examples of co-operative and competitive community behavior among the members of your soil community.
CLEAN UP
1. All culture plates that you are finished with should be discarded in the big orange autoclave bag near the sink next to the instructor table. Ask your instructor whether or not to save stock cultures and plates with organisms that are provided.
2. Culture plates, stocks, etc. that you are not finished with should be labeled on a piece of your your team color tape. Place the labeled cultures in your lab section's designated area in the incubator, the walk-in cold room, or at room temp. in a labeled rack. If you have a stack of plates, wrap a piece of your team color tape around the whole stack.
3. Remove tape from all liquid cultures in glass tubes. Then place the glass tubes with caps in racks by the sink near the instructor's table. Do not discard the contents of the tubes.
4. Glass slides or disposable glass tubes can be discarded in the glass disposal box.
5. Make sure all contaminated, plastic, disposable, serologic pipets and used contaminated micropipet tips are in the small orange autoclave bag sitting in the plastic container on your bench.
6. If you used the microscope, clean the lenses of the microscope with lens paper, being very careful NOT to get oil residue on any of the objectives other than the oil immersion 100x objective. Move the lowest power objective into the locked viewing position, turn off the light source, wind the power cord, and cover the microscope with its dust cover before replacing the microscope in the cabinet.
7. If you used it, rinse your staining tray and leave it upside down on paper towels next to your sink.
8. Turn off the gas and remove the tube from the nozzle. Place your bunsen burner and tube in your large drawer.
9. Place all your equipment (loop, striker, sharpie, etc) including your microfuge rack, your micropipets and your micropipet tips in your small or large drawer.
10. Move your notebook and lab manual so that you can disinfect your bench thoroughly.
11. Take off your lab coat and store it in the blue cabinet with your microscope.
12. Wash your hands.
Assignment
Graded assignment:
Compose a draft of your Materials and Methods section of your final paper with the following general sections: include all work completed through this weeks lab
- Acquiring a soil sample for bacterial community assessment.
- Enumerating bacterial soil community microorganisms.
- Culture-Dependent Microbial Community Assessment of Functional Metabolic Diversity
- Isolation of select soil community bacteria to pure culture and testing Cultured Bacterial Isolates as examples of co-operation and competition among community members.
- Identification of culturable bacteria by 16S rRNA gene sequencing and Maldi-TOF Biotyper
Consult the full directions for this assignment found at Lab 5 assignment: Materials & Methods
Do before next lab:
Characterization of your isolates will start immediately after Spring Break. You will use descendants of the 4-7 isolates amplified today. Because of spring break you need to make sub-cultures of your cultures on FRIDAY March 15 (Tues. & Wed. lab) and by MONDAY March 25 (Tues. Lab) or TUESDAY March 26 (WED. lab). You will also need a fresh broth culture of each of your isolates to start your antibiotic producer testing in Lab 6. Therefore, when you get back from Spring Break, don't forget to come into the lab approximately 24 hours before Lab 6 and set up a new subculture both on a plate and in NA broth using a well isolated colony from each of your last steak plates. You will find a roller in the lab E301 on the bench by the door where you can incubate those broth cultures at room temperature with constant shaking (which will make them grow faster). Follow the directions for making the sub-cultures in broths from a colony on solid medium in the Aseptic Transfer protocol at Aseptic Transfer in Protocols. Please plan ahead to prepare the appropriate cultures so that they will be ready to use when needed. The number of hours it takes from inoculation until a bacterial culture moves from log to stationary or death phase depends on its generation time, the concentration of the inoculum, and other factors.
IN CLASS: Data Analysis Workshop
Soon (in lecture on a day announced by your instructors) you will learn how to use a public data base to analyze your 16s rRNA gene sequences. Please bring your computer, if possible, to that session and make sure you have downloaded any recommended software and signed up for an account on RDP before you come to it. Your instructors will provide information about account opening and downloading software in an announcement in Sakai.
Make sure you have outlined the protocols in your lab notebook and started any necessary cultures on appropriate medium so that they will be ready to use in lab at the appropriate time.
Reference Information
Your most important resource for looking up information about your isolates will be the reference manuals: THE PROKARYOTES and Bergey's Manual. Wellesley College has these valuable reference books available in electronic form. Link to the electronic edition of | The Prokaryotes through Springer ebooks.
Link to the electronic edition of | Bergey's Manualsthrough Springer ebooks.
Use these reference resources to decipher the meaning of the results of the morphologic, metabolic, physical and other tests you will perform or to look up the usual characteristics of the isolates that are identified by 16s rRNA gene sequencing.
Links to Labs
Lab 1
Lab 2
Lab 3
Lab 4
Lab 5
Lab 6
Lab 7
Lab 8
Lab 9
Lab 10
Lab 11