BISC209/S11: Lab2
LAB 2: Identifying and Characterizing Bacteria from a Soil Community
Today you will continue acquiring and practicing skills used by working microbiologists. We will begin our culture-independent approach to identifying bacteria in a soil community by extracting DNA from the quarter gram soil sample that you weighed out last week and your instructor froze for you. The DNA that will be isolated from the soil will be total DNA, a mix of genes and chromosomes from every living thing in that soil sample. Since we are only interested in the bacteria in your soil community, after extracting total (genomic) DNA today, we will freeze it and process it later with the goal of amplifying one bacterial gene of interest that is common to all the bacteria in the soil community but not found in any other type of life form (eukaryotes, archeae, or viruses). We will also continue our culture-dependent approach to isolating and characterizing a few bacteria of interest from our soil sample.
Remember that your semester long project has three parts, two of them are culture based and one is culture-indepedent. We will work on all parts simultaneously. 1) We are in the process (started last week in LAB1) of using traditional culture techniques to isolate and characterize (but not identify) a few of the interesting culturable bacteria in our soil community. You will continue the process of isolation and selection of a few of those bacteria today and for the next several weeks. 2) We will assess a few aspects of cooperative and competitive behavior through culture-dependent community physiological assessment of exoenzyme dependent nutrient processing, and carbon source utilization (beginning in LAB3). The part of our investigation that is culture-independent is to, 3) identify a somewhat random set of bacterial members of your soil community through DNA sequencing of the 16S rRNA gene (rDNA) from soil genomic DNA (beginning today in LAB2).
Tools and Techniques of Microbiologists: Use and Calibration of Micropipets
Because pipetting accurately is crucial to successful quantitative analysis in microbiology, we will begin our work by reviewing proper technique and then practicing the use of micropipets. Your instructor will demonstrate proper technique before you and your partner complete the calibration exercise described below.
How to Use a Micropipettor
The following website Using a Micropipette has more detailed information about micropipette use.
- Decide which of your micropipets is appropriate for the volume you want to measure and dispense. Your P20 can be used for volumes between 2-20μL. If you want to pipet a volume that is less than 2μL you will need to use a P10, P2 or P1 and different tips than those used with the P20 and P200. The P200 can be used for volumes between 21-200μL and the P1000 for volumes between 201-1000μL (1ml). Keep in mind that your P1000 is the least accurate of your pipets;therefore, you may wish to use a 1ml pipet instead.
- Adjust the volume dial to the appropriate volume, recognizing that your P1000 must have a zero added to the bottom of the volume display boxes and the P20 has a decimal between the bottom and second volume display boxes.
- Firmly seat a new micropipet tip of appropriate size on the micropipets.
- Depress with the thumb plunger to the first stop and hold the pipettor in this depressed position (DO NOT depress fully).
- Dip the micropipet into the solution far enough to account for the volume that will be withdrawn but not so far as to immerse the micropipet barrel.
- Gradually release the plunger, drawing fluid into the tip without forming bubbles.
- Carefully slide the micropipet tip along the side of the tube to remove any unwanted droplets of fluid sticking to the tip's surface.
- Expel the fluid into the desired container by touching the micropipet tip to the inside surface of the container and slowly depressing FULLY the plunger.
- Continue to hold the plunger in the fully depressed position as you remove the micropipet from the container.
- Eject the tip by pressing the eject button (if your micropipet has one) into an appropriate place (only tips that have been contaminated with microorganisms need to be ejected into an autoclave bag).
Don'ts in Using Micropipets
- DO NOT force or rotate the volume adjustment knob past the upper or lower ranges specified on the top of the micropipet.
- DO NOT use a micropipet without a tip since the precision piston that measures the volume can be ruined.
- DO NOT put the micropipet down on your bench with a filled tip since fluid can run back into the precision piston.
- DO NOT allow the micropipet plunger to snap back after fluid is either dispensed or drawn.
- DO NOT immerse the micropipet barrel into fluid.
- DO NOT flame the micropipet tip.
Activity: Protocol for Micropipet Calibration
1. To calibrate your P1000, P 200, and P 20 micropipets, label 6 microfuge tubes (1-6) and weigh them on a top loading balance. Don't forget to tare the balance. Record the weights in a table in your lab notebook, like the one below.
2. Using the information in the table below, pipet the specified volumes into the pre-weighed microfuge tubes prepared in step 1 and then reweigh the tubes. Record all weights.
3. Calculate the weight of the water in grams by substracting the dry weight from the weight of the tubes with water. Note that 1000 microliters of water should weigh exactly 1 gram at room temperature.
4. If the water in any tube weighs more or less than 1 gram, repeat that tube. Ask your instructor to check your pipetting technique if your calibration continues to be off after several repeated attempts.
| Tube # | Pre-weight | Tube Vol. in µl using P20 |
Vol. in µl using P200 |
Vol. in µl using P1000 |
Weight of water grams |
|---|---|---|---|---|---|
| 1 | _____ | 10 | 0 | 990 | ____ |
| 2 | _____ | 0 | 100 | 900 | ____ |
| 3 | _____ | 20 | 175 | 805 | _____ |
| 4 | _____ | 2 | 88 | 910 | ______ |
| 5 | _____ | 0 | 200 x 5 | 0 | _____ |
| 6 | _____ | 20 x 5 | 0 | 900 | _____ |
Isolation & Characterization of Cultured Bacteria from a Soil Habitat
Finishing the Standard Plate Count of Culturable Soil Microbial Community
Activity: Last week you started a standard plate count of the culturable microbes in your soil sample. Today you will complete that plate count to get one kind of enumeration of the microorganisms in your soil community. Find a plate that contains 30-300 colonies (there should be only one if you did your 10 fold serial dilution correctly). If it is clear that a culture plate has well over 300 colonies or under 30, designate it as "invalid" in your lab notebook. Count all the surface and subsurface colonies on the selected plate. The colonies can be more easily counted by using a Quebec Colony Counter which allows proper illumination, a grid overlay and by slight magnification of the plate surface. (There are two colony counters in the lab.)
Calculating the number of bacteria per gram of soil
If you divide the number of colonies counted by the amount of inoculum plated times the dilution factor of that plate, you will obtain the number of cultivatable bacteria per gram of soil.
number CFU/(dilution plated*dilution factor) = number of CFU/gram
For example, if you counted 150 colonies on the 10-6 plate the calculation is:
150/(0.1ml plated*1X10-6dilution)= 150X107 which in scientific notation is written as 1.5X109 CFU/gram
Record the number of CFU/ gram of soil in your lab notebook and on the board for comparison with the other sampling sites. This number is colony forming units/ gram WET soil.
Soil bacteria are usually, however, recorded as number of colony forming units in 1 gram of soil DRY weight. Therefore, you will need to weigh each of the three 1 gram samples that you left for oven drying and average the weights. The weights should be considerably less than 1 gram.
Determine the % change in soil weight by subtracting the average dry weight from 1 g wet weight divided by the wet weight, then times 100.
[(wet weight - dry weight)/wet weight] * 100 = % change
Use this % change to convert the CFU per gram wet soil to CFU per gram dry soil. For example: If your dry weight soil averages 0.75 grams, then (1g-0.75g)/1g * (100) = 25% change. The number of microbes should be 25% higher than the number calculated above/gram of wet weight. Calculate 25% (or whatever your conversion factor is) of your CFUs and add that number to the CFUs/ gram of wet weight soil.
Record the number of CFU/ gram DRY weight soil in your lab notebook. If you are unsure of the accuracy of your calculations of CFUs per gram of wet soil weight and per gram of dry weight, check with your instructor. You will need accurate counts to set up community profiling analyses in LAB3.
Things to Consider:
Why did we perform so many dilutions when we set up our plate count in Lab 1?
Why should you only have one plate with 30-300 colonies?
Seal the edges of the nutrient agar plate you counted with parafilm and store the plate in the cold room. You may discard the other plates. If for some reason your enrichment cultures do not provide, eventually, each person on the team with three isolates of the bacteria we seek, you may need to use this plate to find an additional colony of bacteria to isolate in pure culture for study. The bacteria on it should remain viable for a few weeks.
Using Enrichment Media for Isolation and Identification of Soil Bacteria in a Mixed Population
Please watch the YouTube video on streaking for isolation and pay attention to your instructor's demonstration: http://www.youtube.com/watch?v=eyoW18Fzb3o
Directions for Streaking for Isolation are found in the Protocols section of this wiki.
Streaking for Isolation from a Dried Soil Extract:
Each team of students will make a soil extract from one gram of their oven dried soil sample.
Practice Isolation Streaking using Nutrient agar: Nutrient agar is considered "rich" or general purpose medium because it will fulfill the general nutritional needs of most culturable bacteria and its composition will discourage fungal growth (because of the neutral pH). Since using liquid SOIL EXTRACT as your inoculum is analogous to using a broth culture source, you can follow the steps for Broth to Plate transfer found in Aseptic Transfer and in Streaking for Isolation, both found in the Protocols section of this wiki. Aseptic transfer and streaking for isolation are crucially important skills in microbiology. When you are ready to streak out your first plate (after you have read the directions carefully, asked questions to clarify any confusion, and practiced your best technique on an empty plate or piece of paper), call your instructor over and ask her to watch you streak out your first plate. She will give you pointers to enhance your success at repeating this technique on a second plate. Your goal is to have well-separated single colonies when you evaluate your streaked plates next week. Label the plates with your initials, date, lab section, soil sample identifier information, and medium name. Incubate one plate at room temp and one plate at 30C in the racks assigned to your group or Lab section.
Enrichment & Isolation of spore-forming bacteria:
Finding Spore Forming Bacteria
(Such as members of genera Bacillus and of Streptomyces in the Actinomycetes family
Each student will also use a new sterile cotton swab dipped into their oven-dried soil extract to set up an enrichment culture for spore-forming bacteria. Today you will inoculate a plate of glycerol yeast extract agar (GYEA) following the directions for ISOLATION in the Enrichment Media for the Isolation of Soil Bacteria in a Mixed Population: Finding Spore Forming Bacteria in the Protocols: Culture Media section of this wiki.
Isolation of Spore-formers:
Swab section 1 of a labeled plate of glycerol yeast medium (GYEA) using your best isolation streak technique Allow the inoculum in section 1 to absorb into the agar before you switch to your inoculating loop to streak into section 2.
- Follow the steps for Streaking for Isolation in the Protocols section of this wiki. Use your loop to streak sections 2-4.
- Invert, and incubate the plate at RT.
- Check your plate for colonies daily and record your descriptions of the texture and shape of the colonies that appear. If any colonies arise that look tough and leathery or appear as "little, powdered-sugar volcanos", use the tip of a sterile toothpick to pick up a small but visible amount of growth (being careful not to touch anything but the tip of the colony) and "spread" the bacterial growth onto section 1 of a new glycerol yeast plate. (Inoculate one colony/plate.) Use your loop to isolation streak sections 2-4. Don't forget to flame your loop between sections!
- We will select colonies of other potential spore-forming bacteria in lab 3.
Over the next few weeks you will continue to sub-culture onto new plates, using your best isolation streak technique. Your goal is to continue to streak out ONE CFU until you are sure that all the bacterial growth in a colony comes from a single mother cell (pure culture). In subsequent labs you will make a bacterial smear and do a Gram stain of these genetically identical bacteria and you will perform other tests from freshly pure cultures to explore the physical and metabolic characteristics of this isolate.
Actinomycetes and Streptomycetes often form tough leathery colonies, so transfer of these bacteria to new media to start a sub-culture is sometimes difficult. The powdery area may be spores. Take a sample from this area, if possible. In any case, try to "break off" a piece of the colony with your sterile loop or with a sterile toothpick and transfer that whole piece of a colony onto zone one of the new plate. Then use your loop for streaking out the other zones. The tiny spores on the surface of the colony are likely to transfer to the next plate or tube when you work with it. (That's a good thing this time.)
CONTINUE the Enrichment for Hyphomycrobia and Azotobacter from the cultures started last week.
Your team will obtain additional Streaking for Isolation practice by following the directions for SECONDARY ENRICHMENT and ISOLATION using the two enrichment broth cultures that you inoculated last week. This process will continue your attempt to find nitrogen cycling and methylotrophic bacteria. You and your teammates should divide up the work described in the Enrichment Media for the Isolation of Soil Bacteria in a Mixed Population: Finding nitrifying Methylotrophs and Azotobacter.
The directions for Aseptic transfer: Broth to Broth and Broth to Plate are found in the Protocols section of the wiki.
Finding Denitrifying Methylotrophs (Hyphomicrobia) Bacteria :
Use Denitrifying Methylotrophs Medium with and without methanol(DMM)
- Transfer 2 ml of the culture liquid to a fresh screw cap tube of the same DMMM medium with methanol and incubate at 30°C. Incubate for several days. Fill both tubes to the top with DMMM medium in the hood and tighten the screw cap. Place the tubes in the 30°C room. (This is a secondary enrichment.)
- Check for Nitrogen bubbles, as described in Lab 1. If your medium becomes cloudy with bacterial growth and you see the evidence of anaerobic nitrogen gas formation (bubbles), notify your teammates. It is time for each student to isolation streak a plate of DMM (hypomicrobia) solid medium without methanol. We want to end up with each student having a pure culture of an isolate from this secondary enrichment. Incubate the streaked plates of DMM in the closed jar containing an open tube of methanol. The closed container and plates will be kept in the 30°C room. Keep the container in the hood when opening the container to remove or replace a plate or when changing the methanol. Wear gloves and remember that methanol is toxic! Your instructor will change the tube of methanol every few days but if you notice that it is getting low, please remind your instructor to refresh the methanol.
-
Isolation:
After 3 days or so, examine the inoculated DMM plate cultures to look for colonies. Use a dissection microscope or magnifying glass. Look for tiny, glassy (hyaline) colonies, possibly volcano shaped). - Photograph the colonies.
-
Characterization:
- Select a few likely colonies from your original DMM plate and re-streak for isolation onto new DMM (Hyphomicrobia) medium solid medium plates that have NO methanol (one colony/ plate).
- After the bacteria from a single colony are transferred to zone one of a new plate using a sterile toothpick or inoculating needle, you can use your loop to isolation streak out the other zones on each plate. Incubate in the jar (with the open tube of methanol). Check for formation of visible colonies and keep streaking for isolation from a single colony onto fresh plates until you are sure that you have a pure colony isolate. This is particularly difficult with Hyphomicrobiabecause it is easy for other faster growing organisms to overgrow them. When they are in a mixture of bacteria it is particularly difficult to separate them so you will need to do a LOT of sequential subcultures from what appears to be a single colony.)
Hyphomicrobiaalso tend to attach to other bacteria. Reduce the chance of culturing contaminates by transferring newly arising colonies of Hyphomicrobia onto fresh plates as soon as you see them form visible, characteristic colonies. Use your inoculating needle or a sterile toothpick rather than your loop to pick bacteria from the center of your colony to make a new subculture.
Finally, when you feel confident you have a pure isolate of a denitrifying methylotroph (based on colony appearance), do a test to see if these methylotrophic bacteria are actually in the Hyphomicrobiagroup. Perform an isolation streak sub-culture onto nutrient agar. If the bacteria that you transferred grow on NA, they are not Hyphomicrobia. Hyphomicrobia can not grow on nutrient agar; however, don't be upset. You probably have isolated bacteria of a different group of methylotrophic bacteria such as (Pseudomonas spp, Lolium spp, Methylobacterium spp etc.).Continue your characterization of your isolate.
Continue to subculture your isolate onto a fresh plate each week using your best isolation streak technique. Eventually, you will be ready to make a bacterial smear and do a Gram stain and other special stains. We will also perform other tests to explore the physical and metabolic characteristics of this isolate in later labs.
Finding Nitrogen Cyclying Bacteria: Azotobacter
Use Azotobacter Medium
- Secondary enrichment:
-
Each group should take a loopful of the slime from the pellicle (media surface) or from the side of the tube and place it into 1 ml of sterile water in a small tube. Cap the small tube and place the capped tube into an empty 16 mm tube.
- Vortex the big tube to disperse the sample in the small tube. (Vortexing in this way helps break up the other microbes that will be embedded in the slimy material. The other microbes are taking advantage of the by-products of Nitrogen compounds excreted by the N2 fixers. )
- Each student should isolation streak from that tube of vortexed slime onto two plates of Azotobacteria agar medium suspension, using your best isolation streak technique. (Follow the protocol in Streaking for Isolation ).
- Incubate one streak plate at room temp and the other at 30 °C.
-
Isolation:
Watch for the appearance of isolated, slimy colonies on either plate.
Continue to isolation streak to make sub-cultures onto fresh Azotobacter plates until you think you have a pure isolate.
Once you believe you have a pure isolate, continue to subculture it onto a fresh plate each week, using your best isolation streak technique. In a later lab you will make a bacterial smear and do a Gram stain and you will eventually perform other tests to explore its physical and metabolic characteristics.
To differentiate Azotobacter from contaminants: The contaminants colony morphology should appear different from the colony morphology of Azotobacter when streaked onto nutrient agar. Streak a well isolated colony from Azotobacter medium onto a nutrient agar plate and incubate it at room temperature. If more than one type of colony appears on the NA plate, then Gram stain bacteria from each type of colony (keeping track of which slide comes from which colony) to check for morphology, arrangement, size, or other distinguishing differences between contaminates and the Azotobacters you seek. Remember that Azotobacters may not appear slimy on NA so you will have to be careful in deciding which of the different colonies seen on NA contain your Azotobacter bacteria. Restreak the colony with the Azotobacter characteristics onto fresh Azotobacter medium. Characterization:
The complete procedures for Enrichment and Isolation of Azotobacteria and Nitrifying Methylotrophic bacteria are described in Enrichment Media for the Isolation of Soil Bacteria in a Mixed Population: Finding Nitrifying Methylotrophs and Azotobacter protocols).
For more information about culture characteristics of Azotobacter refer to: The Prokaryotes and Bergeys. For an online example of images of colonies and stained cells of Azotobacter vinelandii [1] http://inst.bact.wisc.edu/inst/index.php?module=book&func=displayarticle&art_id=274
Begin Culture Independent Soil Bacteria Community ID by Analysis of 16s rRNA Gene
Isolate Genomic DNA From Soil Sample
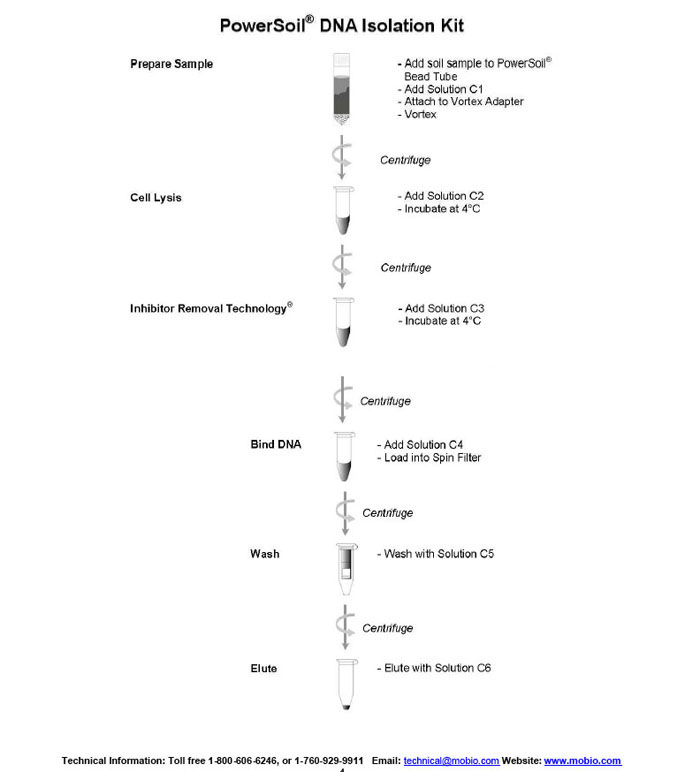
Protocol for Using the Power® Soil DNA Isolation Kit by Mo Bio Laboratories'
Manufacturer Information at [2]
Please wear gloves during this protocol
Activity
1. Each student will do this genomic soil DNA isolation. Be sure to wear gloves throughout this whole procedure to avoid adding skin organisms to your sample. You will each use one 0.25 g soil sample that you weighed out in LAB1 and gave to your instructor for freezing. Transfer ALL of the soil from the microfuge tube to a PowerBead Tube and label this tube on the top only with your initials and a soil sample identifier (A, B, C etc.) on a piece of your team color tape.
What’s happening?: After your sample has been loaded into the PowerBead Tube, the next step is a homogenization and lysis procedure. The PowerBead Tube contains a buffer that will (a) help disperse the soil particles, (b) begin to dissolve humic acids and (c) protect nucleic acids from degradation.
2. Gently vortex to mix.
What’s happening: Gentle vortexing mixes the components in the PowerBead Tube and begins to disperse the sample in the PowerBead Solution.
3. Check Solution C1 to see that it's not precipitated. If Solution C1 is precipitated, heat solution to 60C until the precipitate has dissolved before use.
What’s happening: Solution C1 contains SDS and other disruption agents required for complete cell lysis. In addition to aiding in cell lysis, SDS is an anionic detergent that breaks down fatty acids and lipids associated with the cell membrane of several organisms. If it gets cold, it will form a white precipitate in the bottle. Heating to 60C will dissolve the SDS and will not harm the SDS or the other disruption agents. Solution C1 can be used while it is still warm.
4. Add 60 microliters of Solution C1 to your PowerBead tube and invert several times or vortex briefly.
5. Give your labeled PowerBead tube to your instructor who will take all the PowerBead Tubes to a TOMY® power vortex in the 4°C coldroom. Your samples will "turbovortex" for 7 min at full speed. Alternatively, we can use a FastPrep® Bead Beater for 45 seconds at speed 5 (or full speed. Note: only 2-3 samples will fit in the Fast Prep® BeadBeater at a time.
What’s happening: The BeadBeating step is critical for complete homogenization and cell lysis. Cells are lysed by a combination of chemical agents from steps 1-4 and mechanical shaking introduced at this step. By randomly shaking the beads in the presence of disruption agents, collision of the beads with microbial cells will cause the cells to break open.
6. Microcentrifuge your tubes at 10,000rcf for 1 minute at room temperature. Caution: Be sure not to exceed 10,000rcf or the tubes may break. Make sure the PowerBead tubes rotate freely in your centrifuge without rubbing!
7. Transfer the supernatant (don't transfer the beads!) to a clean 2 ml Collection Tube at the instructor's desk. If you don't know what a collection tube is, ask your instructor. Don't use a regular microfuge tube.
Note: Expect between 400 to 500 microliters of supernatant at this step. The exact recovered volume depends on the absorbancy of your starting material and is not critical for the procedure to be effective. The supernatant may be dark in appearance and still contain some soil particles. The presence of carry over soil or a dark color in the mixture is expected in many soil types at this step.
Subsequent steps in the protocol will remove both carry over soil and coloration of the mixture.
8. Add 250 microliters of Solution C2 to the collection tube and vortex for 5 seconds. Incubate at 4C for 5 minutes.
What’s happening: Solution C2 contains a patented reagent to precipitate non-DNA organic and inorganic material including humic substances, cell debris, and proteins. It is important to remove contaminating organic and inorganic matter that may reduce DNA purity and inhibit downstream DNA applications.
9. Centrifuge the Collection Tube at room temperature for 1 minute at 10,000rcf.
10. Avoiding the pellet, transfer up to, but no more, than 600 microliters of supernatant to a clean 2 ml Collection Tube (provided).
What’s happening: The pellet at this point contains non-DNA organic and inorganic material including humic acid, cell debris, and proteins. For the best DNA yields, and quality, avoid transferring any of the pellet.
11. Add 200 microliters of Solution C3 and vortex briefly. Incubate at 4C for 5 minutes.
What’s happening: Solution C3 is a second reagent (patented) to precipitate additional non-DNA organic and inorganic material including humic acid, cell debris, and proteins. It is important to remove contaminating organic and inorganic matter that may reduce DNA purity and inhibit downstream DNA applications.
12. Centrifuge the tube at room temperature for 1 minute at 10,000rcf.
13. Avoiding the pellet, transfer up to, but no more, than 750 microliters of supernatant to a clean 2 ml Collection Tube (provided).
What’s happening: The pellet at this point contains additional non-DNA organic and inorganic material including humic acid, cell debris, and proteins. For the best DNA yields, and quality, avoid transferring any of the pellet.
14. Shake to mix Solution C4 before use. Add 1.2 ml (do this by adding 600 microliters twice) of Solution C4 to the supernatant (be careful solution doesn’t exceed rim of tube) and vortex for 5 seconds.
What’s happening: Solution C4 has a high concentration of salts. Since DNA binds tightly to silica at high salt concentrations, this will adjust the DNA solution salt concentrations to allow binding of DNA, but not non-DNA organic and inorganic material that may still be present at low levels, to the Spin Filters.
15. Load approximately 675 microliters of the C4 + supernatant mixture from the previous step onto a Spin Filter sitting in a Collection Tube (save the remainder of the supernatant!!) and centrifuge the spin filter at 10,000rcf for 1 minute at room temperature. Discard the flow through (NOT the spin filter!!!) and put the spin filter back in the Collection Tube. Add an additional 675 microliters of the Step 14 mixture to the same Spin Filter and centrifuge at 10,000rcf for 1 minute at room temperature. Discard the flow through and load the remainder of the Step 14 mixture onto the Spin Filter in the Collection Tube and centrifuge at 10,000rcf for 1 minute at room temperature.
Note: A total of three loads for each sample processed are required. You will using the same Spin Filter and Collection Tube for all 3 spins.
What’s happening: DNA is selectively bound to the silica membrane in the Spin Filter device in the high salt solution. Contaminants pass through the filter membrane, leaving only DNA bound to the membrane.
16. Add 500 microliters of Solution C5 to the Spin Filter in the Collection Tube and centrifuge at room temperature for 30 seconds at 10,000rcf.
What’s happening: Solution C5 is an ethanol based wash solution used to further clean the DNA that is bound to the silica filter membrane in the Spin Filter. This wash solution removes residual salt, humic acid, and other contaminants while allowing the DNA to stay bound to the silica membrane.
17. Discard the flow through (not the Spin Filter) from the 2 ml Collection Tube.
What’s happening: This flow through fraction is just non-DNA organic and inorganic waste removed from the silica Spin Filter membrane by the ethanol wash solution.
18. Centrifuge the Spin Filter in the Collection Tube again at room temperature for 1 minute at 10,000rcf.
What’s happening: This second spin removes residual Solution C5 (ethanol wash solution). It is critical to remove all traces of wash solution because the ethanol in Solution C5 can interfere with many downstream DNA applications such as PCR, restriction digests, and gel electrophoresis.
19. Carefully place Spin Filter in a clean 2 ml Collection Tube (provided). DO NOT transfer any liquid that may be on the bottom of the spin filter basket and avoid splashing any Solution C5 onto the Spin Filter.
Note: It is important to avoid any traces of the ethanol based wash solution in the elution that will be created in the next step.
20. Add 100 microliters of Solution C6 to the center of the white Spin Filter membrane.
Note: Placing the Solution C6 (sterile elution buffer) in the center of the small white membrane will make sure the entire membrane is wetted. This will result in a more efficient and complete release of the DNA from the silica Spin Filter membrane. As Solution C6 (elution buffer) passes through the silica membrane, DNA that was bound in the presence of high salt is selectively released by Solution C6 (10 mM Tris) which lacks salt.
Alternatively, sterile DNA-Free PCR Grade Water may be used for DNA elution from the silica Spin Filter membrane at this step (MO BIO Catalog# 17000-10). Solution C6 contains no EDTA. If DNA degradation is a concern, Sterile TE may also be used instead of Solution C6 for elution of DNA from the Spin Filter.
21. Centrifuge the Spin Filter in its Collection Tube at room temperature for 30 seconds at 10,000rcf.
22. Discard the Spin Filter. The DNA in the collection tube is now eluted and ready for freezing or for use as a PCR template.
Storing DNA:
DNA is eluted in Solution C6 (10 mM Tris) and must be used immediately or stored at -20to -80C to prevent degradation.
Make sure your DNA is labeled. We need to measure the DNA concentration before we freeze your extract. DNA at 100ng/microliter is desirable for the next step (amplification of 16s rDNA by polymerase chain reaction) in our culture independent approach to understanding the make up of your soil community. Once you know the concentrate of your DNA, you can adjust the volume of DNA template used in the pcr reaction. The directions for determining the DNA concentration in your extract is described below.
Measuring the Concentration of DNA
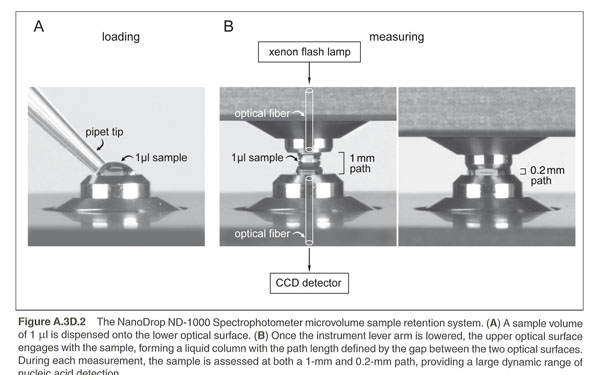
There are two Nanodroppers in the BISC Equipment room, L308. Both the ThermoScientific NanoDrop 2000 and The NanoDrop ND-1000 Spectrophotometers measure DNA by taking Absorbance at A260nm. These spectrophotometers use only 1 microliter of sample and do not require cuvettes. The sample is held in place by fiber optic technology and surface tension that holds the sample in place between two optical surfaces that define the pathlength vertically and dynamically. Measurement can be assessed in a range of 2 to 3700nm/microliter dsDNA. These are expensive machines so make sure you follow the directions carefully and ask your instructor for guidance as needed.
More information is available from the manufacturer's website at: | http://www.nanodrop.com/HowItWorks.aspx
Activity: Using the Nanodroppers
1. Clean the upper and lower optical surfaces of the sample retension device by pipetting 2 microliters of clean deionized water onto the lower optical surface. Close the lever arm and tap it a few times to bathe the upper optical surface. Lift the lever arm and wipe off both optical surfaces with a Kimwipe.

2. Open the NanoDrop software from the Desktop of the computer and select the nucleic acids module.
3. Initialize the machine by placing 1 microliter of clean deionized water onto the lower optic surface, lower the lever arm, and select initialize from the NanoDrop software. Once initialization is complete (~10sec.), clean both optical surfaces with a Kimwipe.

4. Perform a blank measurement by loading 1 microliter of Solution 6 (10mM Tris) and select Blank. Note that this blanking step may use something other than Tris depending on what your sample is dissolve in. Often the blank will be deionized water if you have concentrated your DNA sample already with the ethanol precipation and resolubilized it in water.
Note that as in traditional spectroscopy, the blank will be subtracted from subsequent measurements. If you want to determine the contribution of a specific buffer or diluent, measure the buffer first using distilled water as a blank. If the buffer does not contribute to the A 260nm reading, then deionized water will be fine to use as the blank. The water or buffer should always be measured to be sure that the instrument has been zeroed properly. The measurement of water or buffer should be zero or very close. All measurements are automatically normalized to 340nm.
5. Measure the nucleic acid sample by loading 1microliter of sample and selecting "measure". Record your DNA concentration. Once the measurement is complete. Clean both optical surfaces with a Kimwipe and the machine is ready for the next sample.
You should ensure that the appropriate constant (50 for dsDNA or 40 for RNA) has been chosen. The software automatically calculates the nucleic acid concentration. If the calculation is done by hand, the A260nm is represented as a 1cm path for convenience, even though 1-nm and 0.2nm paths are actually used during the measurement cycle.
Clean Up
When the last sample was been measured, clean the sampling device by repeating step 1.
How to prepare the DNA for pcr amplification of 16S rDNA
The final volume of eluted DNA from the DNA isolation procedure should be ~100μL. We would like for our DNA to be at a concentration of ~ 100ng/μL for our pcr amplification. If your DNA is more concentrated than that, use the "Diluting the DNA" calculations described in the next paragraph to adjust the concentration. If your DNA isolate didn't achieve the concentration we desire, we will adjust our pcr reaction template volume. Directions for doing that can be found in the pcr protocol in Lab 4.
Diluting the DNA
If your isolate is more concentrated than 100ng/μL, you can dilute it in sterile water or 10mM Tris (solution 6). Ask your instructor which is preferred. To calculate how to dilute your DNA, you will use the formula V1 x C1 = V2 X C2, where V1 is ~100μL volume of your eluted DNA from the PowerSoil isolation, C1 is the DNA concentration you obtained from the Nanodroper, C2 is 100ng/μL (the conc. you desire), and V2 is what you will solve for. Please check with your instructor after you have made this calculation and BEFORE you dilute your DNA.
Mobio provides helpful information on [soil DNA isolation issues] http://www.mobio.com/blog/2009/11/08/molecular-biology-of-soil-an-introduction/.
CLEAN UP
1. All culture plates that you are finished with should be discarded in the big orange autoclave bag near the sink next to the instructor table. Ask your instructor whether or not to save stock cultures and plates with organisms that are provided.
2. Culture plates, stocks, etc. that you are not finished with should be labeled on a piece of your your team color tape. Place the labeled cultures in your lab section's designated area in the incubator, the walk-in cold room, or at room temp. in a labeled rack. If you have a stack of plates, wrap a piece of your team color tape around the whole stack.
3. Remove tape from all liquid cultures in glass tubes. Then place the glass tubes with caps in racks by the sink near the instructor's table. Do not discard the contents of the tubes.
4. Glass slides or disposable glass tubes can be discarded in the glass disposal box.
5. Make sure all contaminated, plastic, disposable, serologic pipets and used contaminated micropipet tips are in the small orange autoclave bag sitting in the plastic container on your bench.
6. If you used the microscope, clean the lenses of the microscope with lens paper, being very careful NOT to get oil residue on any of the objectives other than the oil immersion 100x objective. Move the lowest power objective into the locked viewing position, turn off the light source, wind the power cord, and cover the microscope with its dust cover before replacing the microscope in the cabinet.
7. If you used it, rinse your staining tray and leave it upside down on paper towels next to your sink.
8. Turn off the gas and remove the tube from the nozzle. Place your bunsen burner and tube in your large drawer.
9. Place all your equipment (loop, striker, sharpie, etc) including your microfuge rack, your micropipets and your micropipet tips in your small or large drawer.
10. Move your notebook and lab manual so that you can disinfect your bench thoroughly.
11. Take off your lab coat and store it in the blue cabinet with your microscope.
12. Wash your hands.
13. See you next time!
Assignment
Someone from your team must go to the Greenhouse and collect a new soil sample from your sampling site. On the day of lab next week, BEFORE lab begins, stop by the lab to pick up a plastic bag containing materials for collecting a new soil sample from your sampling site in the greenhouse. You will only need enough soil to fill half of a sterile, small 15ml, orange capped, conical tube. Make sure you get the sample for THE SAME sampling site as your original sample. Again avoid the top few millimeters of surface soil and wear gloves to avoid contaminating the sample with bacteria from your skin. You probably won't need to use the corer; a disinfected metal spatula or even a spoon should work fine.
Goal for Cultured Bacteria Isolatiion: Characterize a variety of spore forming, nitrogen cycling, methylotropic, or other interesting bacteria in a soil community
One goal in this project is for your group to end up with 12 unique bacteria (3 each) found in the soil community you sampled. We hope you will have a pure culture from each of the groups described in the Enrichment and Isolation of Soil Bacteria from a Mixed Population, and for you and your partner to have different members of each group. You may not want to limit yourself immediately to attempting to isolate only 3 different colonies. It is not unlikely that some promising bacteria will not survive to the end of the characterization or that several members of the same genus of bacteria are growing on more than one medium, yet may look a bit different on each type. Work with your partners so that you each choose different looking bacterial colonies from each medium and keep consulting with each other to ensure that your subset is unique.
Isolation of desired bacteria from mixed culture is challenging. Your best organizational skills are required. You will be expected to come in at times, on your own, to start, continue or complete this process. We will make every attempt to make the media and reagents that you require available when you need them, but that availability requires advance planning on your part as well as on ours. Communicating your needs or desires well in advance of time of use, will reduce frustration and speed up the process. Remember that because this is an investigative, project based lab course, your instructors do not know the identity of the bacteria you are culturing from your soil habitats. The success of this project is in your hands. Early and continual updating of the plan you devise is crucial. You will not turn in your plan for a grade next week, but your instructor would like for you to create a master plan or flow chart with a preliminary time line and have it in your lab notebook for reference.
Graded Assignment: Make or Fill out a Table of the relevant morphologic, physical, and useful metabolic characteristics of expected genera of soil bacterial that you are attempting to find in your habitat. Be sure to read the directions for this assignment found at: Lab 2 Assignment: Assignment: Table of Cultured Soil Bacteria Characteristics.
Links to Labs
Lab 1
Lab 2
Lab 3
Lab 4
Lab 5
Lab 6
Lab 7
Lab 8
Lab 9
Lab 10
Lab11
Lab 12