Alex A. Cardenas Week 6
From OpenWetWare
Jump to navigationJump to search
Exercise for StARbiochem
- The DNA is the region of molocules that does not enlarge when you switch to the space fill model. The hOGG1 protein is the part of the AA sequence that enlarges when you switch to the space filled model.
- [Cys]241:ASG #2406. The sulfer atom is located in the side chain of the amino acid.
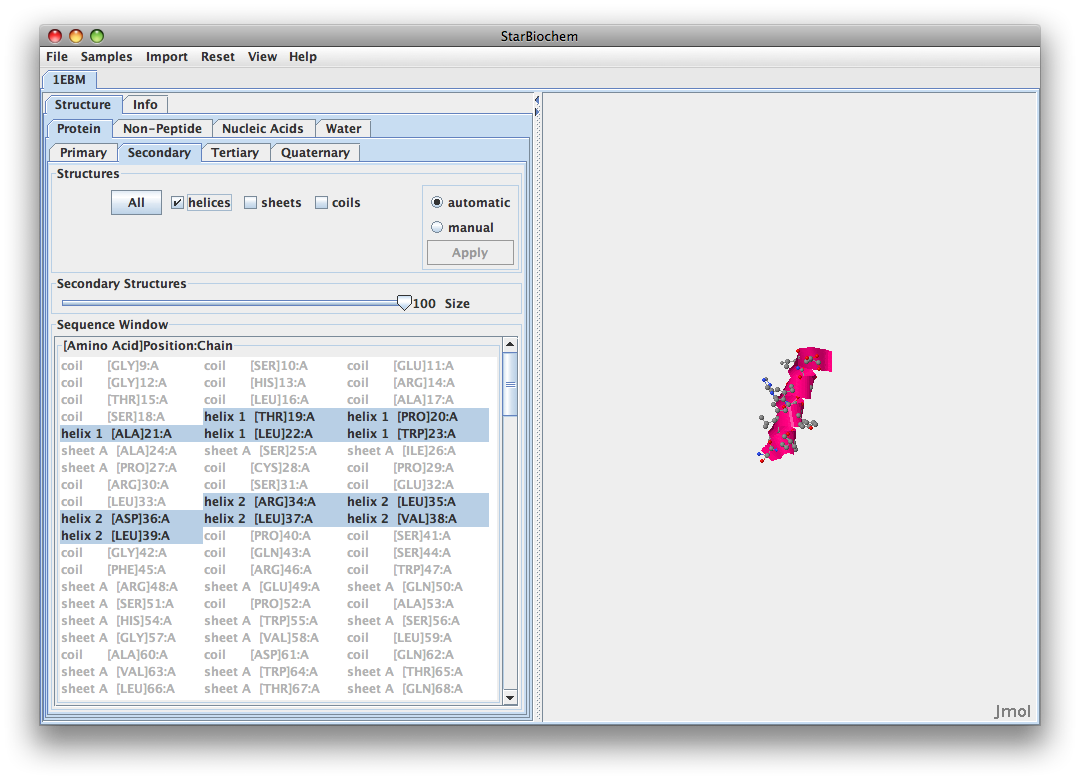
105 THR 106 LEU 107 ALA 108 GLN 109 LEU 110 TYR 111 HIS 112 HIS 113 TRP 114 GLY 115 SER 116 VAL 117 ASP
- Yes there are helicies (pink), sheets (yellow), and coils (blue). The amino acids 105-117 fold into helices. Here is the image
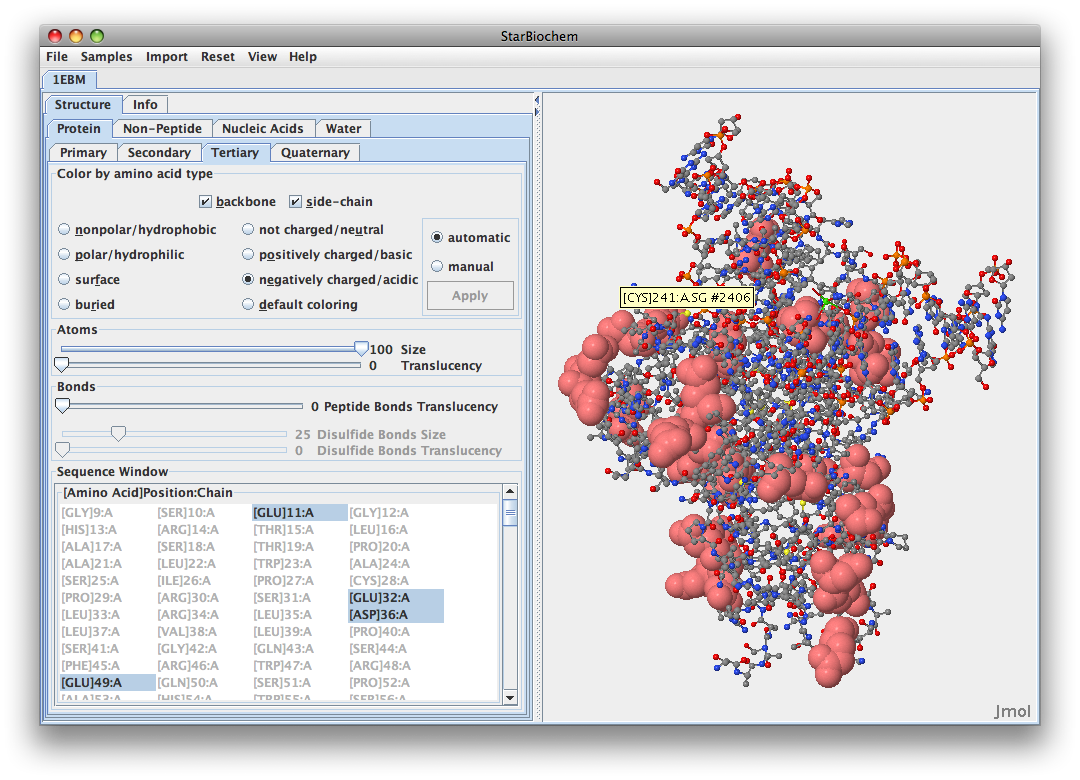
- I would say that the negatively charged amino acids are located on the outside of this protein. This would suggest that the cellular enviornment surrounding this protein is hydrophillic because it would be interacting with the cellular enviorment. I would also say that the hydrophobic regions could be found burried inside the protein. Here is the image:
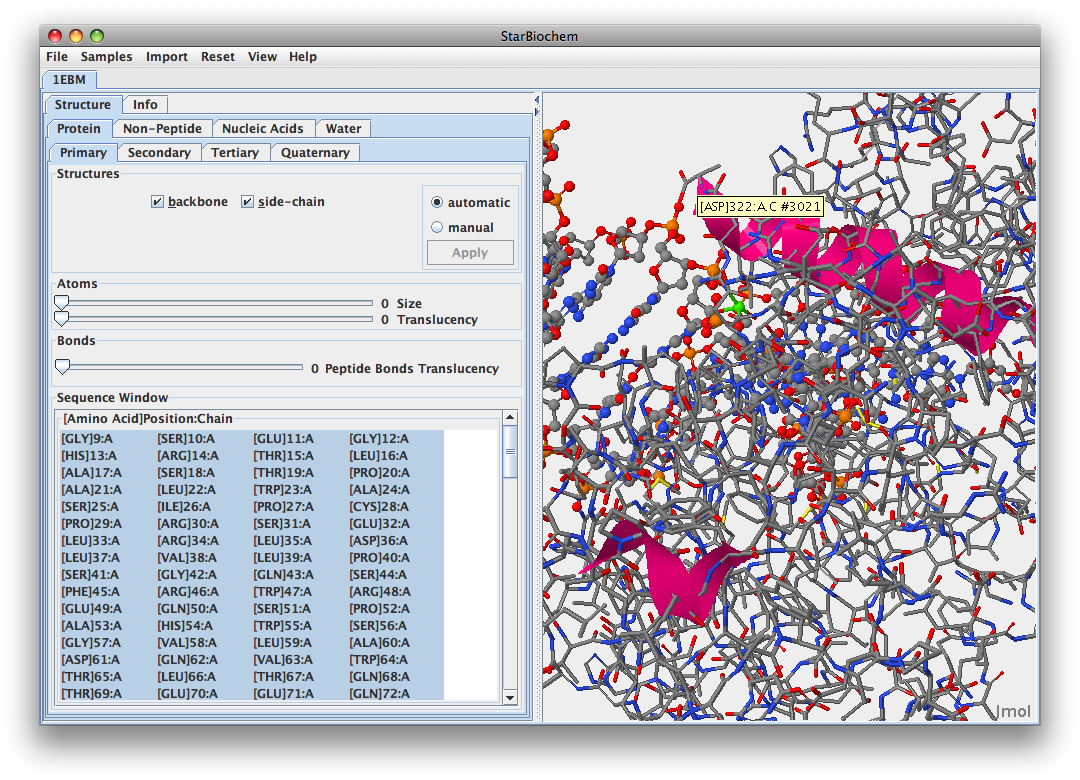 .
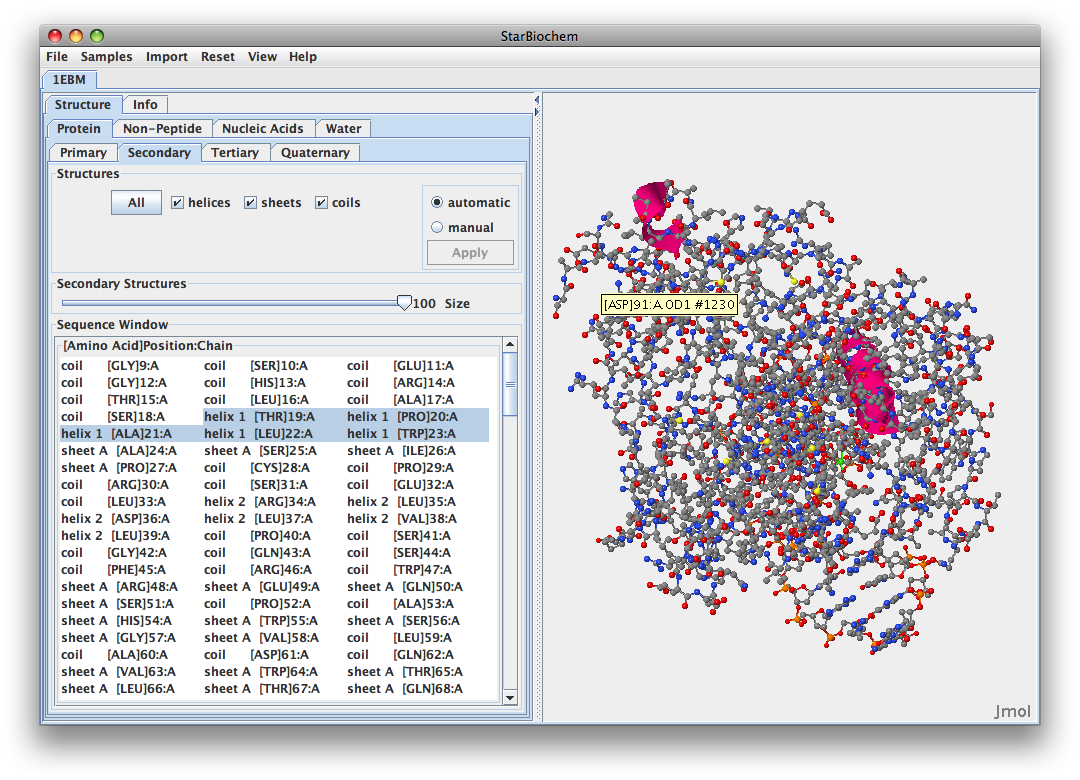
.- You are more likely to find the amino acids that recognize damaged guanine bases withthin hOGG1 in helix 16 because it is closer to the DNA sequence. This is the imaage
 .
.
Exercise for Cn3D
- This is a quaternary protein structure because you can see two polypeptides and the way in which they are going.
- The DNA glycosylase has 3 domains as seen in this image
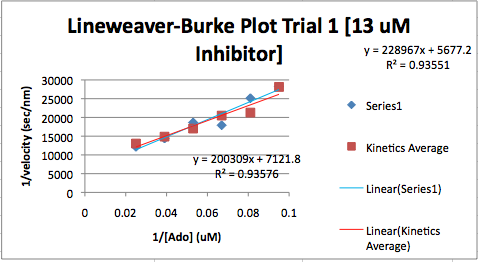
- There are some things you can view the same but I feel that Star Biochem has a lot more options to choose from when looking at so many different things. There are many different options you can choose from in Star Biochem which makes the program really useful.
- Screenshot that is similar to a view of previous screenshot:
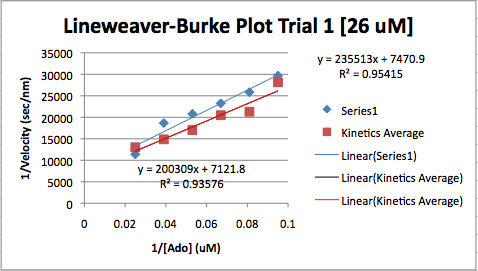
- The program that I prefer is Star Biochem. There are a wide array of different options to choose from including the different amino acids that you would like to have shown, You could look at the protein structure in 4 different ways, each way having multiple subsections to choose from.
Links
- Alex A. Cardenas
- Week 6 Assignment
- Alex A. Cardenas Week 2
- Alex A. Cardenas Week 3
- Alex A. Cardenas Week 4
- Alex A. Cardenas Week 5
- Alex A. Cardenas Week 6
- Alex A. Cardenas Week 7
- Alex A. Cardenas Week 8
- Alex A. Cardenas Week 9
- Alex A. Cardenas Week 10
- Alex A. Cardenas Week 11
- Alex A. Cardenas Week 12
- Alex A. Cardenas Week 13
- Alex A. Cardenas Week 14
- Class Journal Week 1
- Class Journal Week 2
- Class Journal Week 3
- Class Journal Week 5
- Class Journal Week 6
- Class Journal Week 7
- Class Journal Week 8
- Class Journal Week 9
- Class Journal Week 10
- Class Journal Week 11
- Class Journal Week 12
- Class Journal Week 13
- Class Journal Week 14