User:Mbennie/Notebook/Lab Notebook/Notebook/2007/08/09
From OpenWetWare
- Oligos
- IgAbfull-F
- Resuspended at 50uM
- Colony PCR
- Performed on seven colonies of 8.3.2007 IgAbc-R plate
- Template: 20ul Supermix, .8ul VF2 and VR, 1 ul of cell dilution (colony in 100ul of water)
- Plated picked colonies on Amp/Cl plate and grew up at 37C all day and overnight
- Note: Annealing temp was initially 45C, noticed after 5 cycles, restarted protocol without initial 95C denature and correct annealing temperature
- Protocol:
- 95C for 15 mins
- 94C for 30 secs
- 55C for 30 secs
- 68C for 1.5 mins
- REPEAT 2-4 39 times
- 68C for 10 mins
- 4C FOREVER
- PCR
- Full Iga beta: IgAbfull-F and IgAb-R
- Template: 40ul PCR Supermix, .4ul of each primer, and .4ul N. gonorrhea DNA
- Same protocol as the colony PCR
- Digest
- Template (20ul rxns): 3ul DNA (of each), 2ul NEB4, .5ul Sap1, rest water
- C + D with B
- B + C
- C + D
- Thermocycler protocol: 1hr@37C, 20mins@65C
- Template (20ul rxns): 3ul DNA (of each), 2ul NEB4, .5ul Sap1, rest water
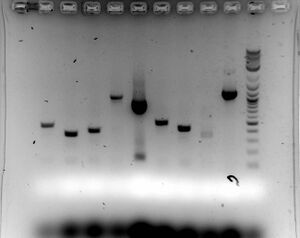
- Gel
- Ran 1.5% for 50 minutes at 100V to see if PCR product is correct length
- Liquid Culture
- Grew up cultures in 8ml Amp/Cl LB
- GCN4 #1 from 8.4 colony PCR backup plate
- GCN4 #3 from 8.4 colony PCR backup plate
- IgAbc #8 from 8.9 colony PCR cell dilution
- Grew up cultures in 8ml Amp/Cl LB
- Ligation
- Performed on today's digest tubes
- Template (50ul rxns): 20ul digest, 5ul ligase buffer, .5ul ligase, rest water
- Thermocycler protocol: 1hr@16C,10mins@65C